Gene conversion and purifying selection shape nucleotide variation in gibbon L/M opsin genes
- PMID: 22017819
- PMCID: PMC3213168
- DOI: 10.1186/1471-2148-11-312
Gene conversion and purifying selection shape nucleotide variation in gibbon L/M opsin genes
Abstract
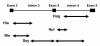
Background: Routine trichromatic color vision is a characteristic feature of catarrhines (humans, apes and Old World monkeys). This is enabled by L and M opsin genes arrayed on the X chromosome and an autosomal S opsin gene. In non-human catarrhines, genetic variation affecting the color vision phenotype is reported to be absent or rare in both L and M opsin genes, despite the suggestion that gene conversion has homogenized the two genes. However, nucleotide variation of both introns and exons among catarrhines has only been examined in detail for the L opsin gene of humans and chimpanzees. In the present study, we examined the nucleotide variation of gibbon (Catarrhini, Hylobatidae) L and M opsin genes. Specifically, we focused on the 3.6~3.9-kb region that encompasses the centrally located exon 3 through exon 5, which encode the amino acid sites functional for the spectral tuning of the genes.
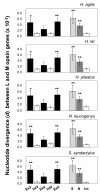
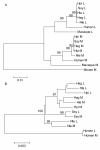
Results: Among 152 individuals representing three genera (Hylobates, Nomascus and Symphalangus), all had both L and M opsin genes and no L/M hybrid genes. Among 94 individuals subjected to the detailed DNA sequencing, the nucleotide divergence between L and M opsin genes in the exons was significantly higher than the divergence in introns in each species. The ratio of the inter-LM divergence to the intra-L/M polymorphism was significantly lower in the introns than that in synonymous sites. When we reconstructed the phylogenetic tree using the exon sequences, the L/M gene duplication was placed in the common ancestor of catarrhines, whereas when intron sequences were used, the gene duplications appeared multiple times in different species. Using the GENECONV program, we also detected that tracts of gene conversions between L and M opsin genes occurred mostly within the intron regions.
Conclusions: These results indicate the historical accumulation of gene conversions between L and M opsin genes in the introns in gibbons. Our study provides further support for the homogenizing role of gene conversion between the L and M opsin genes and for the purifying selection against such homogenization in the central exons to maintain the spectral difference between L and M opsins in non-human catarrhines.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

