Genetic manipulation of genes and cells in the nervous system of the fruit fly
- PMID: 22017985
- PMCID: PMC3232021
- DOI: 10.1016/j.neuron.2011.09.021
Genetic manipulation of genes and cells in the nervous system of the fruit fly
Abstract
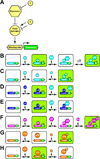
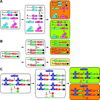
Research in the fruit fly Drosophila melanogaster has led to insights in neural development, axon guidance, ion channel function, synaptic transmission, learning and memory, diurnal rhythmicity, and neural disease that have had broad implications for neuroscience. Drosophila is currently the eukaryotic model organism that permits the most sophisticated in vivo manipulations to address the function of neurons and neuronally expressed genes. Here, we summarize many of the techniques that help assess the role of specific neurons by labeling, removing, or altering their activity. We also survey genetic manipulations to identify and characterize neural genes by mutation, overexpression, and protein labeling. Here, we attempt to acquaint the reader with available options and contexts to apply these methods.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures






References
-
- Adams MD, Sekelsky JJ. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat. Rev. Genet. 2002;3:189–198. - PubMed
-
- Akemann W, Mutoh H, Perron A, Rossier J, Knopfel T. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat. Methods. 2010;7:643–649. - PubMed
-
- Alderson T. Chemically induced delayed germinal mutation in Drosophila. Nature. 1965;207:164–167. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

