A putative vesicular transporter expressed in Drosophila mushroom bodies that mediates sexual behavior may define a neurotransmitter system
- PMID: 22017990
- PMCID: PMC3201771
- DOI: 10.1016/j.neuron.2011.08.032
A putative vesicular transporter expressed in Drosophila mushroom bodies that mediates sexual behavior may define a neurotransmitter system
Abstract
Vesicular transporters are required for the storage of all classical and amino acid neurotransmitters in synaptic vesicles. Some neurons lack known vesicular transporters, suggesting additional neurotransmitter systems remain unidentified. Insect mushroom bodies (MBs) are critical for several behaviors, including learning, but the neurotransmitters released by the intrinsic Kenyon cells (KCs) remain unknown. Likewise, KCs do not express a known vesicular transporter. We report the identification of a novel Drosophila gene portabella (prt) that is structurally similar to known vesicular transporters. Both larval and adult brains express PRT in the KCs of the MBs. Additional PRT cells project to the central complex and optic ganglia. prt mutation causes an olfactory learning deficit and an unusual defect in the male's position during copulation that is rescued by expression in KCs. Because prt is expressed in neurons that lack other known vesicular transporters or neurotransmitters, it may define a previously unknown neurotransmitter system responsible for sexual behavior and a component of olfactory learning.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


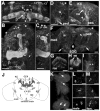

 ). The numbers 4 and 2 indicate primers used to detect the deletion. The lines below show deficiencies that uncover prt (Df(3R)mbc-30, Df(3R)Exel6195). The shaded boxes represent the deletion. The cross-hatched boxes indicate breakpoints approximated by polytene chromosome squash. Scale bars: 50 kb for deficiencies and 0.2 kb for inset. B) PCR was used to detect the prt1 mutation. Wild-type CS flies show a major band at 1.2 kb, prt1 flies show a major band at 400 bp, and prt1/+ heterozygotes show both. (−) indicates a no-template control. A 1 kb standard is shown on the left. C) Single confocal slices through the mushroom body lobes of whole-mount brains labeled with anti-PRT. A w1118 brain is on top with labeling of the MB lobes (α, α′, β, β′, and γ) and heel (h) and a prt1 brain is on bottom, showing no detectable anti-PRT labeling. Scale bar: 20 μm. D) H&E stained paraffin sections show that prt1 mutants have grossly intact MB morphology (α, α′, β, and γ lobes and heel (h), peduncle (ped), and calyx (ca)) and central complex (fan-shaped body (fsb), noduli (nod) and protocerebral bridge (pb)). The β′ lobe is also grossly intact in prt1 (not shown). Scale bar: 20 μm. E, F) Volumetric analyses of the MB calyx (E) and CCX (F) in CS and prt1 did not significantly differ (Student’s t-test, p = 0.1934 (E) and p = 0.2496 (F)). Bars represent means ± SEMs, n = 10 for all groups.
). The numbers 4 and 2 indicate primers used to detect the deletion. The lines below show deficiencies that uncover prt (Df(3R)mbc-30, Df(3R)Exel6195). The shaded boxes represent the deletion. The cross-hatched boxes indicate breakpoints approximated by polytene chromosome squash. Scale bars: 50 kb for deficiencies and 0.2 kb for inset. B) PCR was used to detect the prt1 mutation. Wild-type CS flies show a major band at 1.2 kb, prt1 flies show a major band at 400 bp, and prt1/+ heterozygotes show both. (−) indicates a no-template control. A 1 kb standard is shown on the left. C) Single confocal slices through the mushroom body lobes of whole-mount brains labeled with anti-PRT. A w1118 brain is on top with labeling of the MB lobes (α, α′, β, β′, and γ) and heel (h) and a prt1 brain is on bottom, showing no detectable anti-PRT labeling. Scale bar: 20 μm. D) H&E stained paraffin sections show that prt1 mutants have grossly intact MB morphology (α, α′, β, and γ lobes and heel (h), peduncle (ped), and calyx (ca)) and central complex (fan-shaped body (fsb), noduli (nod) and protocerebral bridge (pb)). The β′ lobe is also grossly intact in prt1 (not shown). Scale bar: 20 μm. E, F) Volumetric analyses of the MB calyx (E) and CCX (F) in CS and prt1 did not significantly differ (Student’s t-test, p = 0.1934 (E) and p = 0.2496 (F)). Bars represent means ± SEMs, n = 10 for all groups.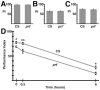

References
-
- Acebes A, Cobb M, Ferveur JF. Species-specific effects of single sensillum ablation on mating position in Drosophila. J Exp Biol. 2003;206:3095–3100. - PubMed
-
- Ackerman SL, Siegel RW. Chemically reinforced conditioned courtship in Drosophila: responses of wild-type and the dunce, amnesiac and don giovanni mutants. J Neurogenet. 1986;3:111–123. - PubMed
-
- Baier A, Wittek B, Brembs B. Drosophila as a new model organism for the neurobiology of aggression? J Exp Biol. 2002;205:1233–1240. - PubMed
-
- Bao X, Wang B, Zhang J, Yan T, Yang W, Jiao F, Liu J, Wang S. Localization of serotonin/tryptophan-hydroxylase-immunoreactive cells in the brain and suboesophageal ganglion of Drosophila melanogaster. Cell Tissue Res. 2010;340:51–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

