High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing
- PMID: 22018332
- PMCID: PMC3333774
- DOI: 10.1186/gb-2011-12-10-r104
High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing
Abstract
RNA interference (RNAi) screening is a state-of-the-art technology that enables the dissection of biological processes and disease-related phenotypes. The commercial availability of genome-wide, short hairpin RNA (shRNA) libraries has fueled interest in this area but the generation and analysis of these complex data remain a challenge. Here, we describe complete experimental protocols and novel open source computational methodologies, shALIGN and shRNAseq, that allow RNAi screens to be rapidly deconvoluted using next generation sequencing. Our computational pipeline offers efficient screen analysis and the flexibility and scalability to quickly incorporate future developments in shRNA library technology.
Figures


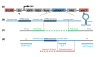


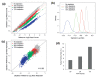

References
-
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, Westbrook TF, Liang AC, Chang K, Hackett JA, Harper JW, Hannon GJ, Elledge SJ. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

