Outcome of experimental porcine circovirus type 1 infections in mid-gestational porcine foetuses
- PMID: 22018436
- PMCID: PMC3216242
- DOI: 10.1186/1746-6148-7-64
Outcome of experimental porcine circovirus type 1 infections in mid-gestational porcine foetuses
Abstract
Background: Porcine circovirus type 1 (PCV1) has been described as a non-cytopathic contaminant of the PK-15 cell line. Several experimental infections with PCV1 failed to reproduce disease in pigs. Therefore, PCV1 is generally accepted as non-pathogenic to pigs. To our knowledge, nothing is known about the outcome of PCV1 infections in porcine foetuses. This was examined in the present study.
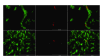
Results: Nine foetuses from three sows were inoculated at 55 days of gestation: three with 10(4.3) TCID(50) of the PCV1 cell culture strain ATCC-CCL33, three with 10(4.3) TCID(50) of the PCV1 field strain 3384 and three with cell culture medium (mock-inoculated). At 21 days post-inoculation, all 6 PCV1-inoculated and all 3 mock-inoculated foetuses had a normal external appearance. Microscopic lesions characterized by severe haemorrhages were observed in the lungs of two foetuses inoculated with CCL33. High PCV1 titres (up to 10(4.7) TCID(50)/g tissue) were found in the lungs of the CCL33-inoculated foetuses. All other organs of the CCL33-inoculated foetuses and all the organs of the 3384-inoculated foetuses were negative (< 10(1.7) TCID(50)/g tissue) by virus titration. PCV1-positive cells (up to 121 cells/10 mm(2) in CCL33-inoculated foetuses and up to 13 cells/10 mm(2) in 3384-inoculated foetuses) were found in the heart, lungs, spleen, liver, thymus and tonsils. PCR and DNA sequencing of Rep recovered CCL33 or 3384 sequences from CCL33- or 3384-inoculated foetuses, respectively.
Conclusions: From this study, it can be concluded that cell culture PCV1 can replicate efficiently and produce pathology in the lungs of porcine foetuses inoculated at 55 days of foetal life.
Figures


References
-
- Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Orig A. 1974;226:153–167. - PubMed
-
- Fenaux M, Halbur PG, Gill M, Toth TE, Meng XJ. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J Clin Microbiol. 2000;38:2494–2503. - PMC - PubMed
-
- Puvanendiran S, Stone S, Yu W, Johnson CR, Abrahante J, Jimenez LG, Griggs T, Haley C, Wagner B, Murtaugh MP. Absence of porcine circovirus type 1 (PCV1) and high prevalence of PCV2 exposure and infection in swine finisher herds. Virus Research. 2011;157:92–98. doi: 10.1016/j.virusres.2011.02.012. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

