Clathrin-mediated endocytosis in budding yeast
- PMID: 22018597
- PMCID: PMC3253927
- DOI: 10.1016/j.tcb.2011.09.001
Clathrin-mediated endocytosis in budding yeast
Abstract
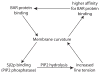
Clathrin-mediated endocytosis in the budding yeast Saccharomyces cerevisiae involves the ordered recruitment, activity and disassembly of nearly 60 proteins at distinct sites on the plasma membrane. Two-color live-cell fluorescence microscopy has proven to be invaluable for in vivo analysis of endocytic proteins: identifying new components, determining the order of protein arrival and dissociation, and revealing even very subtle mutant phenotypes. Yeast genetics and functional genomics facilitate identification of complex interaction networks between endocytic proteins and their regulators. Quantitative datasets produced by these various analyses have made theoretical modeling possible. Here, we discuss recent findings on budding yeast endocytosis that have advanced our knowledge of how -60 endocytic proteins are recruited, perform their functions, are regulated by lipid and protein modifications, and are disassembled, all with remarkable regularity.
Published by Elsevier Ltd.
Figures




References
-
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115(4):475–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

