Stepwise expansion of the bacteriophage ϕ6 procapsid: possible packaging intermediates
- PMID: 22019738
- PMCID: PMC3223026
- DOI: 10.1016/j.jmb.2011.10.004
Stepwise expansion of the bacteriophage ϕ6 procapsid: possible packaging intermediates
Abstract
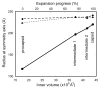
The initial assembly product of bacteriophage ϕ6, the procapsid, undergoes major structural transformation during the sequential packaging of its three segments of single-stranded RNA. The procapsid, a compact icosahedrally symmetric particle with deeply recessed vertices, expands to the spherical mature capsid, increasing the volume available to accommodate the genome by 2.5-fold. It has been proposed that expansion and packaging are linked, with each stage in expansion presenting a binding site for a particular RNA segment. To investigate procapsid transformability, we induced expansion by acidification, heating, and elevated salt concentration. Cryo-electron microscopy reconstructions after all three treatments yielded the same partially expanded particle. Analysis by cryo-electron tomography showed that all vertices of a given capsid were either in a compact or an expanded state, indicating a highly cooperative transition. To benchmark the mature capsid, we analyzed filled (in vivo packaged) capsids. When these particles were induced to release their RNA, they reverted to the same intermediate state as expanded procapsids (intermediate 1) or to a second, further expanded state (intermediate 2). This partial reversibility of expansion suggests that the mature spherical capsid conformation is obtained only when sufficient outward pressure is exerted by packaged RNA. The observation of two intermediates is consistent with the proposed three-step packaging process. The model is further supported by the observation that a mutant capable of packaging the second RNA segment without previously packaging the first segment has enhanced susceptibility for switching spontaneously from the procapsid to the first intermediate state.
Published by Elsevier Ltd.
Figures





References
-
- Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 2004;101:83–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

