Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster
- PMID: 22019781
- PMCID: PMC3576853
- DOI: 10.1038/ng.948
Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster
Abstract
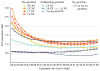
Many animal species use a chromosome-based mechanism of sex determination, which has led to the coordinate evolution of dosage-compensation systems. Dosage compensation not only corrects the imbalance in the number of X chromosomes between the sexes but also is hypothesized to correct dosage imbalance within cells that is due to monoallelic X-linked expression and biallelic autosomal expression, by upregulating X-linked genes twofold (termed 'Ohno's hypothesis'). Although this hypothesis is well supported by expression analyses of individual X-linked genes and by microarray-based transcriptome analyses, it was challenged by a recent study using RNA sequencing and proteomics. We obtained new, independent RNA-seq data, measured RNA polymerase distribution and reanalyzed published expression data in mammals, C. elegans and Drosophila. Our analyses, which take into account the skewed gene content of the X chromosome, support the hypothesis of upregulation of expressed X-linked genes to balance expression of the genome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: biological implications. Trends Genet. 2005;21:219–226. - PubMed
-
- Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. - PubMed
-
- Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

