Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes
- PMID: 22019887
- PMCID: PMC3553601
- DOI: 10.1038/nm.2454
Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes
Abstract
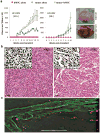
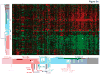
Development and preclinical testing of new cancer therapies is limited by the scarcity of in vivo models that authentically reproduce tumor growth and metastatic progression. We report new models for breast tumor growth and metastasis in the form of transplantable tumors derived directly from individuals undergoing treatment for breast cancer. These tumor grafts illustrate the diversity of human breast cancer and maintain essential features of the original tumors, including metastasis to specific sites. Co-engraftment of primary human mesenchymal stem cells maintains phenotypic stability of the grafts and increases tumor growth by promoting angiogenesis. We also report that tumor engraftment is a prognostic indicator of disease outcome for women with newly diagnosed breast cancer; orthotopic breast tumor grafting is a step toward individualized models for tumor growth, metastasis and prognosis. This bank of tumor grafts also serves as a publicly available resource for new models in which to study the biology of breast cancer.
Figures





References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

