A scalable pipeline for highly effective genetic modification of a malaria parasite
- PMID: 22020067
- PMCID: PMC3431185
- DOI: 10.1038/nmeth.1742
A scalable pipeline for highly effective genetic modification of a malaria parasite
Abstract
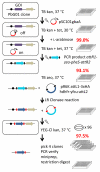
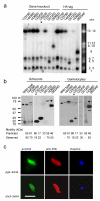
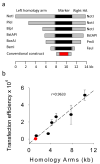
In malaria parasites, the systematic experimental validation of drug and vaccine targets by reverse genetics is constrained by the inefficiency of homologous recombination and by the difficulty of manipulating adenine and thymine (A+T)-rich DNA of most Plasmodium species in Escherichia coli. We overcame these roadblocks by creating a high-integrity library of Plasmodium berghei genomic DNA (>77% A+T content) in a bacteriophage N15-based vector that can be modified efficiently using the lambda Red method of recombineering. We built a pipeline for generating P. berghei genetic modification vectors at genome scale in serial liquid cultures on 96-well plates. Vectors have long homology arms, which increase recombination frequency up to tenfold over conventional designs. The feasibility of efficient genetic modification at scale will stimulate collaborative, genome-wide knockout and tagging programs for P. berghei.
Figures


References
METHODS – ADDITIONAL REFERENCES
-
- Janse CJ, Franke-Fayard B, Waters AP. Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat Protoc. 2006;1:614–623. doi:10.1038/nprot.2006.88. - PubMed
-
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. - PubMed
References
-
- van Dijk MR, Janse CJ, Waters AP. Expression of a Plasmodium gene introduced into subtelomeric regions of Plasmodium berghei chromosomes. Science. 1996;271:662–665. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

