The retinoid X receptors and their ligands
- PMID: 22020178
- PMCID: PMC4097889
- DOI: 10.1016/j.bbalip.2011.09.014
The retinoid X receptors and their ligands
Abstract
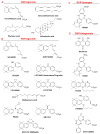
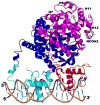
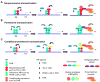
This chapter presents an overview of the current status of studies on the structural and molecular biology of the retinoid X receptor subtypes α, β, and γ (RXRs, NR2B1-3), their nuclear and cytoplasmic functions, post-transcriptional processing, and recently reported ligands. Points of interest are the different changes in the ligand-binding pocket induced by variously shaped agonists, the communication of the ligand-bound pocket with the coactivator binding surface and the heterodimerization interface, and recently identified ligands that are natural products, those that function as environmental toxins or drugs that had been originally designed to interact with other targets, as well as those that were deliberately designed as RXR-selective transcriptional agonists, synergists, or antagonists. Of these synthetic ligands, the general trend in design appears to be away from fully aromatic rigid structures to those containing partial elements of the flexible tetraene side chain of 9-cis-retinoic acid. This article is part of a Special Issue entitled Advances in High Density Lipoprotein Formation and Metabolism: A Tribute to John F. Oram (1945-2010).
© 2011 Elsevier B.V. All rights reserved.
Figures


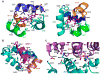

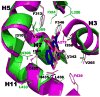
References
-
- Wolf G. Is 9-cis-retinoic acid the endogenous ligand for the retinoic acid-X receptor? Nutr Rev. 2006;64:532–538. - PubMed
-
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. - PubMed
-
- Feige JN, Gelman L, Tudor C, Engelborghs Y, Wahli W, Desvergne B. Fluorescence imaging reveals the nuclear behavior of peroxisome proliferator-activated receptor/retinoid X receptor heterodimers in the absence and presence of ligand. J Biol Chem. 2005;280:17880–17890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

