ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding
- PMID: 22020299
- PMCID: PMC3210321
- DOI: 10.1038/nsmb.2135
ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding
Abstract
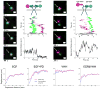
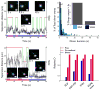
The extent to which ligand occupancy and dimerization contribute to erbB1 signaling is controversial. To examine this, we used two-color quantum-dot tracking for visualization of the homodimerization of human erbB1 and quantification of the dimer off-rate (k(off)) on living cells. Kinetic parameters were extracted using a three-state hidden Markov model to identify transition rates between free, co-confined and dimerized states. We report that dimers composed of two ligand-bound receptors are long-lived and their k(off) is independent of kinase activity. By comparison, unliganded dimers have a more than four times faster k(off). Transient co-confinement of receptors promotes repeated encounters and enhances dimer formation. Mobility decreases more than six times when ligand-bound receptors dimerize. Blockade of erbB1 kinase activity or disruption of actin networks results in faster diffusion of receptor dimers. These results implicate both signal propagation and the cortical cytoskeleton in reduced mobility of signaling-competent erbB1 dimers.
Conflict of interest statement
No conflicts of interest are declared.
Figures



Comment in
-
Finding the missing links in EGFR.Nat Struct Mol Biol. 2012 Jan 5;19(1):1-3. doi: 10.1038/nsmb.2221. Nat Struct Mol Biol. 2012. PMID: 22218287 No abstract available.
References
-
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. - PubMed
-
- Ferguson KM, et al. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. - PubMed
-
- Chung I, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. - PubMed
-
- Clayton AH, et al. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

