Experimental evolution of a facultative thermophile from a mesophilic ancestor
- PMID: 22020511
- PMCID: PMC3255606
- DOI: 10.1128/AEM.05773-11
Experimental evolution of a facultative thermophile from a mesophilic ancestor
Abstract
Experimental evolution via continuous culture is a powerful approach to the alteration of complex phenotypes, such as optimal/maximal growth temperatures. The benefit of this approach is that phenotypic selection is tied to growth rate, allowing the production of optimized strains. Herein, we demonstrate the use of a recently described long-term culture apparatus called the Evolugator for the generation of a thermophilic descendant from a mesophilic ancestor (Escherichia coli MG1655). In addition, we used whole-genome sequencing of sequentially isolated strains throughout the thermal adaptation process to characterize the evolutionary history of the resultant genotype, identifying 31 genetic alterations that may contribute to thermotolerance, although some of these mutations may be adaptive for off-target environmental parameters, such as rich medium. We undertook preliminary phenotypic analysis of mutations identified in the glpF and fabA genes. Deletion of glpF in a mesophilic wild-type background conferred significantly improved growth rates in the 43-to-48°C temperature range and altered optimal growth temperature from 37°C to 43°C. In addition, transforming our evolved thermotolerant strain (EVG1064) with a wild-type allele of glpF reduced fitness at high temperatures. On the other hand, the mutation in fabA predictably increased the degree of saturation in membrane lipids, which is a known adaptation to elevated temperature. However, transforming EVG1064 with a wild-type fabA allele had only modest effects on fitness at intermediate temperatures. The Evolugator is fully automated and demonstrates the potential to accelerate the selection for complex traits by experimental evolution and significantly decrease development time for new industrial strains.
Figures


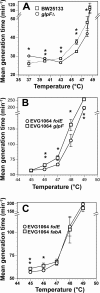
References
-
- Angelov A, Mientus M, Liebl S, Liebl W. 2009. A two-host fosmid system for functional screening of (meta)genomic libraries from extreme thermophiles. Syst. Appl. Microbiol. 32:177–185 - PubMed
-
- Antal M, Bordeau V, Douchin V, Felden B. 2005. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 280:7901–7908 - PubMed
-
- Barrick JE, et al. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

