Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7
- PMID: 22020516
- PMCID: PMC3255626
- DOI: 10.1128/AEM.06231-11
Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7
Abstract
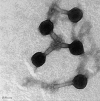
Salmonella enterica and Escherichia coli O157:H7 are major food-borne pathogens causing serious illness. Phage SFP10, which revealed effective infection of both S. enterica and E. coli O157:H7, was isolated and characterized. SFP10 contains a 158-kb double-stranded DNA genome belonging to the Vi01 phage-like family Myoviridae. In vitro adsorption assays showed that the adsorption constant rates to both Salmonella enterica serovar Typhimurium and E. coli O157:H7 were 2.50 × 10⁻⁸ ml/min and 1.91 × 10⁻⁸ ml/min, respectively. One-step growth analysis revealed that SFP10 has a shorter latent period (25 min) and a larger burst size (>200 PFU) than ordinary Myoviridae phages, suggesting effective host infection and lytic activity. However, differential development of resistance to SFP10 in S. Typhimurium and E. coli O157:H7 was observed; bacteriophage-insensitive mutant (BIM) frequencies of 1.19 × 10⁻² CFU/ml for S. Typhimurium and 4.58 × 10⁻⁵ CFU/ml for E. coli O157:H7 were found, indicating that SFP10 should be active and stable for control of E. coli O157:H7 with minimal emergence of SFP10-resistant pathogens but may not be for S. Typhimurium. Specific mutation of rfaL in S. Typhimurium and E. coli O157:H7 revealed the O antigen as an SFP10 receptor for both bacteria. Genome sequence analysis of SFP10 and its comparative analysis with homologous Salmonella Vi01 and Shigella phiSboM-AG3 phages revealed that their tail fiber and tail spike genes share low sequence identity, implying that the genes are major host specificity determinants. This is the first report identifying specific infection and inhibition of Salmonella Typhimurium and E. coli O157:H7 by a single bacteriophage.
Figures






References
-
- Altermann E, Klaenhammer TR. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7:161–169. - PubMed
-
- Barbara G, et al. 2000. Role of antibiotic therapy on long-term germ excretion in faeces and digestive symptoms after Salmonella infection. Aliment. Pharmacol. Ther. 14:1127–1131 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

