The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308
- PMID: 22020650
- PMCID: PMC3256608
- DOI: 10.1128/JB.05623-11
The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308
Abstract
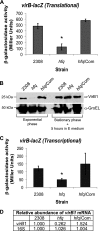
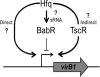
The type IV secretion system encoded by the virB operon is required for full virulence of Brucella sp., and the present study links the RNA chaperone Hfq to wild-type expression of virB in Brucella abortus 2308. Studies employing virB-lacZ fusions, quantitative reverse transcription-PCR, and immunoblot analysis showed that both transcription and translation of virB are decreased in an isogenic hfq mutant compared to those in the parental strain. These results led to the hypothesis that Hfq regulation of virB is mediated through an intermediate transcriptional regulator. Subsequent experiments determined that expression of the gene encoding the putative Brucella quorum-sensing regulator BabR (also known as BlxR), a known virB regulator, is also controlled by Hfq at the posttranscriptional level, and a cis-acting element in the 5' untranslated region of the babR transcript responsible for this regulation was identified. Consistent with its role as a virB regulator, recombinant Brucella BabR binds to the virB promoter region in electrophoretic mobility shift assays. However, experiments employing a babR mutant strain determined that BabR is a repressor, not an activator, of virB transcription. These findings suggest that Hfq regulates virB expression through both BabR-dependent and BabR-independent pathways.
Figures


References
-
- Aiba H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10:134–139 - PubMed
-
- Celli J, Grovel JP. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 7:93–97 - PubMed
-
- Comerci DJ, Martínez-Lorenzo MJ, Sieira R, Grovel JP, Ugalde RA. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159–168 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

