Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system
- PMID: 22021388
- PMCID: PMC3249375
- DOI: 10.1534/genetics.111.132779
Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system
Abstract
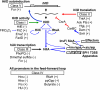
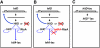
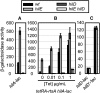
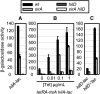
Salmonella enterica serovar Typhimurium uses the Salmonella pathogenicity island 1 (SPI1) type III secretion system to induce inflammatory diarrhea and bacterial uptake into intestinal epithelial cells. The expression of hilA, encoding the transcriptional activator of the SPI1 structural genes, is directly controlled by three AraC-like regulators, HilD, HilC, and RtsA, each of which can activate the hilD, hilC, rtsA, and hilA genes, forming a complex feed-forward regulatory loop. A large number of factors and environmental signals have been implicated in SPI1 regulation. We have developed a series of genetic tests that allows us to determine where these factors feed into the SPI1 regulatory circuit. Using this approach, we have grouped 21 of the known SPI1 regulators and environmental signals into distinct classes on the basis of observed regulatory patterns, anchored by those few systems where the mechanism of regulation is best understood. Many of these factors are shown to work post-transcriptionally at the level of HilD, while others act at the hilA promoter or affect all SPI1 promoters. Analysis of the published transcriptomic data reveals apparent coregulation of the SPI1 and flagellar genes in various conditions. However, we show that in most cases, the factors that affect both systems control SPI1 independently of the flagellar protein FliZ, despite its role as an important SPI1 regulator and coordinator of the two systems. These results provide a comprehensive model for SPI1 regulation that serves as a framework for future molecular analyses of this complex regulatory network.
Figures
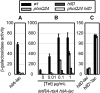
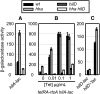
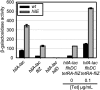
References
-
- Bailly-Bechet M., Benecke A., Hardt W. D., Lanza V., Sturm A., et al. , 2010. An externally modulated, noise-driven switch for the regulation of SPI1 in Salmonella enterica serovar Typhimurium. J. Math. Biol. PMID: 21107576 - PubMed
-
- Bajaj V., Hwang C., Lee C. A., 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18: 715–727 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

