A sibling-augmented case-only approach for assessing multiplicative gene-environment interactions
- PMID: 22021562
- PMCID: PMC3246688
- DOI: 10.1093/aje/kwr231
A sibling-augmented case-only approach for assessing multiplicative gene-environment interactions
Abstract
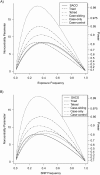
Family-based designs protect analyses of genetic effects from bias that is due to population stratification. Investigators have assumed that this robustness extends to assessments of gene-environment interaction. Unfortunately, this assumption fails for the common scenario in which the genotyped variant is related to risk through linkage with a causative allele. Bias also plagues other methods of assessment of gene-environment interaction. When testing against multiplicative joint effects, the case-only design offers excellent power, but it is invalid if genotype and exposure are correlated in the population. The authors describe 4 mechanisms that produce genotype-exposure dependence: exposure-related genetic population stratification, effects of family history on behavior, genotype effects on exposure, and selective attrition. They propose a sibling-augmented case-only (SACO) design that protects against the former 2 mechanisms and is therefore valid for studying young-onset disease in which genotype does not influence exposure. A SACO design allows the ascertainment of genotype and exposure for cases and exposure for 1 or more unaffected siblings selected randomly. Conditional logistic regression permits assessment of exposure effects and gene-environment interactions. Via simulations, the authors compare the likelihood-based inference on interactions using the SACO design with that based on other designs. They also show that robust analyses of interactions using tetrads or disease-discordant sibling pairs are equivalent to analyses using the SACO design.
Figures
References
-
- Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13(2):153–162. - PubMed
-
- Chatterjee N, Kalaylioglu Z, Carroll RJ. Exploiting gene-environment independence in family-based case-control studies: increased power for detecting associations, interactions and joint effects. Genet Epidemiol. 2005;28(2):138–156. - PubMed
-
- Self SG, Longton G, Kopecky KJ, et al. On estimating HLA/disease association with application to a study of aplastic anemia. Biometrics. 1991;47(1):53–61. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

