TOX is required for development of the CD4 T cell lineage gene program
- PMID: 22021617
- PMCID: PMC3221881
- DOI: 10.4049/jimmunol.1101474
TOX is required for development of the CD4 T cell lineage gene program
Abstract
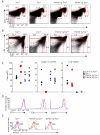
The factors that regulate thymic development of the CD4(+) T cell gene program remain poorly defined. The transcriptional regulator ThPOK is a dominant factor in CD4(+) T cell development, which functions primarily to repress the CD8 lineage fate. Previously, we showed that nuclear protein TOX is also required for murine CD4(+) T cell development. In this study, we sought to investigate whether the requirement for TOX was solely due to a role in ThPOK induction. In apparent support of this proposition, ThPOK upregulation and CD8 lineage repression were compromised in the absence of TOX, and enforced ThPOK expression could restore some CD4 development. However, these "rescued" CD4 cells were defective in many aspects of the CD4(+) T cell gene program, including expression of Id2, Foxo1, and endogenous Thpok, among others. Thus, TOX is necessary to establish the CD4(+) T cell lineage gene program, independent of its influence on ThPOK expression.
Figures





References
-
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

