Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium
- PMID: 22021638
- PMCID: PMC3243482
- DOI: 10.1194/jlr.M020313
Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium
Abstract
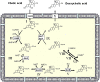
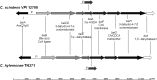
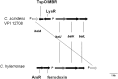
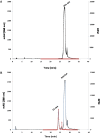
The human bile acid pool composition is composed of both primary bile acids (cholic acid and chenodeoxycholic acid) and secondary bile acids (deoxycholic acid and lithocholic acid). Secondary bile acids are formed by the 7α-dehydroxylation of primary bile acids carried out by intestinal anaerobic bacteria. We have previously described a multistep biochemical pathway in Clostridium scindens that is responsible for bile acid 7α-dehydroxylation. We have identified a large (12 kb) bile acid inducible (bai) operon in this bacterium that encodes eight genes involved in bile acid 7α-dehydroxylation. However, the function of the baiF gene product in this operon has not been elucidated. In the current study, we cloned and expressed the baiF gene in E. coli and discovered it has bile acid CoA transferase activity. In addition, we discovered a second bai operon encoding three genes. The baiK gene in this operon was expressed in E. coli and found to encode a second bile acid CoA transferase. Both bile acid CoA transferases were determined to be members of the type III family by amino acid sequence comparisons. Both bile acid CoA transferases had broad substrate specificity, except the baiK gene product, which failed to use lithocholyl-CoA as a CoA donor. Primary bile acids are ligated to CoA via an ATP-dependent mechanism during the initial steps of 7α-dehydroxylation. The bile acid CoA transferases conserve the thioester bond energy, saving the cell ATP molecules during bile acid 7α-dehydroxylation. ATP-dependent CoA ligation is likely quickly supplanted by ATP-independent CoA transfer.
Figures



References
-
- Ridlon J. M., Kang D., Hylemon P. B. 2006. Bile salt biotransformations by Human Intestinal bacteria. J. Lipid Res. 47: 241–259. - PubMed
-
- Bernstein H., Bernstein C., Payne C. M., Dvorakova K., Garewal H. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589: 47–65. - PubMed
-
- Berr F., Kullak-Ublick G. A., Paumgartner G., Munzig W., Hylemon P. B. 1996. 7alpha-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology. 111: 1611–1620. - PubMed
-
- Dowling R. H., Veysey M. J., Pereira S. P., Hussaini S. H., Thomas L. A., Wass J. A., Murphy G. M. 1997. Role of intestinal transit in the pathogenesis of gallbladder stones. Can. J. Gastroenterol. 11: 57–64. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

