The dynamics of supply and demand in mRNA translation
- PMID: 22022250
- PMCID: PMC3192816
- DOI: 10.1371/journal.pcbi.1002203
The dynamics of supply and demand in mRNA translation
Abstract
We study the elongation stage of mRNA translation in eukaryotes and find that, in contrast to the assumptions of previous models, both the supply and the demand for tRNA resources are important for determining elongation rates. We find that increasing the initiation rate of translation can lead to the depletion of some species of aa-tRNA, which in turn can lead to slow codons and queueing. Particularly striking "competition" effects are observed in simulations of multiple species of mRNA which are reliant on the same pool of tRNA resources. These simulations are based on a recent model of elongation which we use to study the translation of mRNA sequences from the Saccharomyces cerevisiae genome. This model includes the dynamics of the use and recharging of amino acid tRNA complexes, and we show via Monte Carlo simulation that this has a dramatic effect on the protein production behaviour of the system.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
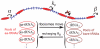
 , a
, a  aa-tRNA is removed from the pool, and a
aa-tRNA is removed from the pool, and a  tRNA is added to the corresponding pool of bare tRNAs. Bare
tRNA is added to the corresponding pool of bare tRNAs. Bare  tRNAs are recharged with a rate
tRNAs are recharged with a rate  .
.
 for each type of tRNA used in the simulations. These are based on the gene copy number for each tRNA in the Saccharomyces cerevisiae genome. The key for the label
for each type of tRNA used in the simulations. These are based on the gene copy number for each tRNA in the Saccharomyces cerevisiae genome. The key for the label  of each codon is available in the supporting information (Text S2).
of each codon is available in the supporting information (Text S2).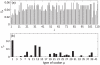
 of the codon at each site and (b) the occurrence frequency
of the codon at each site and (b) the occurrence frequency  of each codon type on mRNA A.
of each codon type on mRNA A.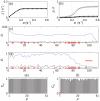
 , respectively. In (b) the points show the reader density
, respectively. In (b) the points show the reader density  and the crosses the coverage density
and the crosses the coverage density  . Plots (c) and (d) show ribosome density as a function of position
. Plots (c) and (d) show ribosome density as a function of position  for small (
for small ( ) and large (
) and large ( ) initiation rate respectively. Black lines show the reader density
) initiation rate respectively. Black lines show the reader density  and blue lines the coverage density
and blue lines the coverage density  . Red dots show the positions of codons of type
. Red dots show the positions of codons of type  , and the red bar indicates the width of the ribosomes. Bar graphs (e) and (f) show the steady state charging rate
, and the red bar indicates the width of the ribosomes. Bar graphs (e) and (f) show the steady state charging rate  of each tRNA type. (e) shows
of each tRNA type. (e) shows  and (f)
and (f)  , the same values as in (c) and (d).
, the same values as in (c) and (d).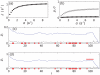
 and the crosses the coverage density
and the crosses the coverage density  . Plots (c) and (d) show ribosome density as a function of position
. Plots (c) and (d) show ribosome density as a function of position  for small (
for small ( ) and large (
) and large ( ) initiation rate respectively. Black lines show the reader density
) initiation rate respectively. Black lines show the reader density  and blue lines the coverage density
and blue lines the coverage density  . Red dots show codons of type
. Red dots show codons of type  as in Fig. 4.
as in Fig. 4.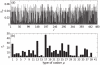
 of the codon at each site and (b) the occurrence frequency
of the codon at each site and (b) the occurrence frequency  of each codon type on mRNA B.
of each codon type on mRNA B.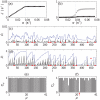
 ) and large (
) and large ( ) initiation rate respectively. Red dots show the positions of codons of type
) initiation rate respectively. Red dots show the positions of codons of type  , and the red bar indicates the width of the ribosomes. (e) and (f) show
, and the red bar indicates the width of the ribosomes. (e) and (f) show  for
for  the same as in (c) and (d).
the same as in (c) and (d).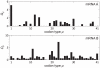
 for each codon type for mRNAs A and B.
for each codon type for mRNAs A and B.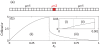
 . Which kind of queueing will be observed depends on
. Which kind of queueing will be observed depends on  , and the three regimes discussed in the text are separated with dotted lines. The inset shows a zoom around small
, and the three regimes discussed in the text are separated with dotted lines. The inset shows a zoom around small  .
.
 and
and  as a function of
as a function of  for mRNAs of type A (black points) and type B (red crosses) (the same initiation rates are used for each species). The blue line shows
for mRNAs of type A (black points) and type B (red crosses) (the same initiation rates are used for each species). The blue line shows  , where blue labelled codons cause queueing. Plot (c) shows the charging levels of tRNAs for
, where blue labelled codons cause queueing. Plot (c) shows the charging levels of tRNAs for  . (d) and (e) show the site dependent reader (pale lines) and coverage (dark lines) density for each mRNA type, again for
. (d) and (e) show the site dependent reader (pale lines) and coverage (dark lines) density for each mRNA type, again for  . The codons corresponding to the first aa-tRNA to become depleted are highlighted with blue dots (
. The codons corresponding to the first aa-tRNA to become depleted are highlighted with blue dots ( ), and those for the second in green (
), and those for the second in green ( ). Plots (f)–(j) show similar results for a mixture in the ratio 80∶20; results for a 20∶80 mixture are presented in the supporting information (Text S4).
). Plots (f)–(j) show similar results for a mixture in the ratio 80∶20; results for a 20∶80 mixture are presented in the supporting information (Text S4).
 of the first two aa-tRNAs to become depleted. Plot (a) shows results for the 20∶80 mixture of mRNAs A and B, plot (b) the 50∶50 mixture, and (c) the 80∶20 mixture. From left to right the abundance of mRNA A increases. Blue and green lines correspond to the codons labelled blue and green in Fig. 10, and dashed lines show
of the first two aa-tRNAs to become depleted. Plot (a) shows results for the 20∶80 mixture of mRNAs A and B, plot (b) the 50∶50 mixture, and (c) the 80∶20 mixture. From left to right the abundance of mRNA A increases. Blue and green lines correspond to the codons labelled blue and green in Fig. 10, and dashed lines show  .
.
 and
and  as a function of
as a function of  for mRNAs of type C (black points) and type D (red crosses). The blue line shows
for mRNAs of type C (black points) and type D (red crosses). The blue line shows  , where blue labelled codons first become depleted. Plot (c) shows the charging levels of tRNAs for
, where blue labelled codons first become depleted. Plot (c) shows the charging levels of tRNAs for  . (d) and (e) show the site dependent reader (pale lines) and coverage (dark lines) density for each mRNA type. The codons corresponding to the first aa-tRNA to become depleted are highlighted with blue dots (
. (d) and (e) show the site dependent reader (pale lines) and coverage (dark lines) density for each mRNA type. The codons corresponding to the first aa-tRNA to become depleted are highlighted with blue dots ( ), and those for the second in green (
), and those for the second in green ( ). Plots (f)–(j) show similar results for a mixture in the ratio 20∶80; results for a 80∶20 mixture are presented in the supporting information (Text S4).
). Plots (f)–(j) show similar results for a mixture in the ratio 20∶80; results for a 80∶20 mixture are presented in the supporting information (Text S4).
 of the first two aa-tRNAs to become depleted. Plot (a) shows results for the 20∶50 mixture of mRNAs C and D, plot (b) the 50∶50 mixture, and (c) the 80∶20 mixture. From left to right the abundance of mRNA C increases. In each case the tRNA type labelled blue becomes depleted first. The inset in (c) is a zoom at small
of the first two aa-tRNAs to become depleted. Plot (a) shows results for the 20∶50 mixture of mRNAs C and D, plot (b) the 50∶50 mixture, and (c) the 80∶20 mixture. From left to right the abundance of mRNA C increases. In each case the tRNA type labelled blue becomes depleted first. The inset in (c) is a zoom at small  showing this more clearly. Dashed lines show
showing this more clearly. Dashed lines show  .
.
References
-
- Day D, Tuite M. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol. 1998;157:361–371. - PubMed
-
- Sørensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in escherichia coli. J Mol Biol. 1989;207:365–377. - PubMed
-
- Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective Charging of tRNA Isoacceptors Explains Patterns of Codon Usage. Science. 2003;300:1718–1722. - PubMed
-
- Alberts B, Johnson A, Walter P, Lewis J. Molecular Biology of the Cell. Garland Pub. Inc, 5th edition; 2008.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

