The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile
- PMID: 22022270
- PMCID: PMC3192846
- DOI: 10.1371/journal.ppat.1002317
The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile
Abstract
Nosocomial infections are increasingly being recognised as a major patient safety issue. The modern hospital environment and associated health care practices have provided a niche for the rapid evolution of microbial pathogens that are well adapted to surviving and proliferating in this setting, after which they can infect susceptible patients. This is clearly the case for bacterial pathogens such as Methicillin Resistant Staphylococcus aureus (MRSA) and Vancomycin Resistant Enterococcus (VRE) species, both of which have acquired resistance to antimicrobial agents as well as enhanced survival and virulence properties that present serious therapeutic dilemmas for treating physicians. It has recently become apparent that the spore-forming bacterium Clostridium difficile also falls within this category. Since 2000, there has been a striking increase in C. difficile nosocomial infections worldwide, predominantly due to the emergence of epidemic or hypervirulent isolates that appear to possess extended antibiotic resistance and virulence properties. Various hypotheses have been proposed for the emergence of these strains, and for their persistence and increased virulence, but supportive experimental data are lacking. Here we describe a genetic approach using isogenic strains to identify a factor linked to the development of hypervirulence in C. difficile. This study provides evidence that a naturally occurring mutation in a negative regulator of toxin production, the anti-sigma factor TcdC, is an important factor in the development of hypervirulence in epidemic C. difficile isolates, presumably because the mutation leads to significantly increased toxin production, a contentious hypothesis until now. These results have important implications for C. difficile pathogenesis and virulence since they suggest that strains carrying a similar mutation have the inherent potential to develop a hypervirulent phenotype.
Conflict of interest statement
The authors have declared that no competing interests exist.
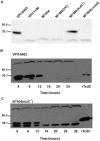
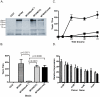
Figures

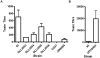
References
-
- Borriello SP. Pathogenesis of Clostridium difficile infection. J Antimicrob Chemother. 1998;41:13–19. - PubMed
-
- Warny M, Pepin J, Fang A, Killgore G, Thompson A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–1084. - PubMed
-
- McDonald LC, Killgore GE, Thompson A, Owens RC, Kazakova SV, et al. An epidemic, toxin gene–variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–2441. - PubMed
-
- Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–2449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

