Murine gammaretrovirus group G3 was not found in Swedish patients with myalgic encephalomyelitis/chronic fatigue syndrome and fibromyalgia
- PMID: 22022360
- PMCID: PMC3192035
- DOI: 10.1371/journal.pone.0024602
Murine gammaretrovirus group G3 was not found in Swedish patients with myalgic encephalomyelitis/chronic fatigue syndrome and fibromyalgia
Abstract
Background: The recent report of gammaretroviruses of probable murine origin in humans, called xenotropic murine retrovirus related virus (XMRV) and human murine leukemia virus related virus (HMRV), necessitated a bioinformatic search for this virus in genomes of the mouse and other vertebrates, and by PCR in humans.
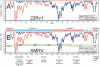
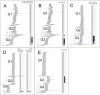
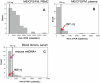
Results: Three major groups of murine endogenous gammaretroviruses were identified. The third group encompassed both exogenous and endogenous Murine Leukemia Viruses (MLVs), and most XMRV/HMRV sequences reported from patients suffering from myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Two sensitive real-time PCRs for this group were developed. The predicted and observed amplification range for these and three published XMRV/HMRV PCRs demonstrated conspicuous differences between some of them, partly explainable by a recombinatorial origin of XMRV. Three reverse transcription real-time PCRs (RTQPCRs), directed against conserved and not overlapping stretches of env, gag and integrase (INT) sequences of XMRV/HMRV were used on human samples. White blood cells from 78 patients suffering from ME/CFS, of which 30 patients also fulfilled the diagnostic criteria for fibromyalgia (ME/CFS/FM) and in 7 patients with fibromyalgia (FM) only, all from the Gothenburg area of Sweden. As controls we analyzed 168 sera from Uppsala blood donors. We controlled for presence and amplifiability of nucleic acid and for mouse DNA contamination. To score as positive, a sample had to react with several of the XMRV/HMRV PCRs. None of the samples gave PCR reactions which fulfilled the positivity criteria.
Conclusions: XMRV/HMRV like proviruses occur in the third murine gammaretrovirus group, characterized here. PCRs developed by us, and others, approximately cover this group, except for the INT RTQPCR, which is rather strictly XMRV specific. Using such PCRs, XMRV/HMRV could not be detected in PBMC and plasma samples from Swedish patients suffering from ME/CFS/FM, and in sera from Swedish blood donors.
Conflict of interest statement
Figures



References
-
- Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585–589. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

