Detection of a fourth orbivirus non-structural protein
- PMID: 22022432
- PMCID: PMC3192121
- DOI: 10.1371/journal.pone.0025697
Detection of a fourth orbivirus non-structural protein
Abstract
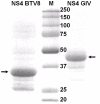
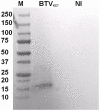
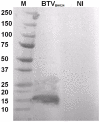
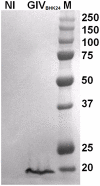
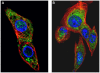
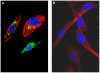
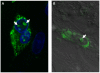
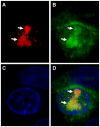
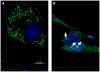
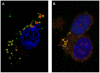
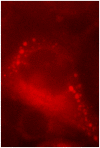
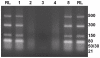
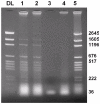
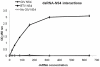
The genus Orbivirus includes both insect and tick-borne viruses. The orbivirus genome, composed of 10 segments of dsRNA, encodes 7 structural proteins (VP1-VP7) and 3 non-structural proteins (NS1-NS3). An open reading frame (ORF) that spans almost the entire length of genome segment-9 (Seg-9) encodes VP6 (the viral helicase). However, bioinformatic analysis recently identified an overlapping ORF (ORFX) in Seg-9. We show that ORFX encodes a new non-structural protein, identified here as NS4. Western blotting and confocal fluorescence microscopy, using antibodies raised against recombinant NS4 from Bluetongue virus (BTV, which is insect-borne), or Great Island virus (GIV, which is tick-borne), demonstrate that these proteins are synthesised in BTV or GIV infected mammalian cells, respectively. BTV NS4 is also expressed in Culicoides insect cells. NS4 forms aggregates throughout the cytoplasm as well as in the nucleus, consistent with identification of nuclear localisation signals within the NS4 sequence. Bioinformatic analyses indicate that NS4 contains coiled-coils, is related to proteins that bind nucleic acids, or are associated with membranes and shows similarities to nucleolar protein UTP20 (a processome subunit). Recombinant NS4 of GIV protects dsRNA from degradation by endoribonucleases of the RNAse III family, indicating that it interacts with dsRNA. However, BTV NS4, which is only half the putative size of the GIV NS4, did not protect dsRNA from RNAse III cleavage. NS4 of both GIV and BTV protect DNA from degradation by DNAse. NS4 was found to associate with lipid droplets in cells infected with BTV or GIV or transfected with a plasmid expressing NS4.
Conflict of interest statement
Figures









References
-
- Mertens PP, Attoui H, Duncan R, Dermody TS. Reoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, A BL, editors. Virus Taxonomy Eighth Report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2005. pp. 447–454.
-
- Sangar DV, Mertens PP. Comparison of type 1 bluetongue virus protein synthesis in vivo and in vitro. In: Compans RW, Bishop DHL, editors. Double-Stranded RNA Viruses. New York: Elsevier; 1983. pp. 183–191.
-
- Grubman MJ, Appleton JA, Letchworth GJ., 3rd Identification of bluetongue virus type 17 genome segments coding for polypeptides associated with virus neutralization and intergroup reactivity. Virology. 1983;131:355–366. - PubMed
-
- Mertens PP, Brown F, Sangar DV. Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virology. 1984;135:207–217. - PubMed
-
- Wade-Evans AM, Mertens PP, Belsham GJ. Sequence of genome segment 9 of bluetongue virus (serotype 1, South Africa) and expression analysis demonstrating that different forms of VP6 are derived from initiation of protein synthesis at two distinct sites. J Gen Virol. 1992;73 (Pt 11):3023–3026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

