Infection of XC cells by MLVs and Ebola virus is endosome-dependent but acidification-independent
- PMID: 22022555
- PMCID: PMC3192169
- DOI: 10.1371/journal.pone.0026180
Infection of XC cells by MLVs and Ebola virus is endosome-dependent but acidification-independent
Abstract
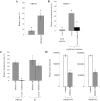
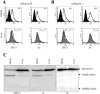
Inhibitors of endosome acidification or cathepsin proteases attenuated infections mediated by envelope proteins of xenotropic murine leukemia virus-related virus (XMRV) and Ebola virus, as well as ecotropic, amphotropic, polytropic, and xenotropic murine leukemia viruses (MLVs), indicating that infections by these viruses occur through acidic endosomes and require cathepsin proteases in the susceptible cells such as TE671 cells. However, as previously shown, the endosome acidification inhibitors did not inhibit these viral infections in XC cells. It is generally accepted that the ecotropic MLV infection in XC cells occurs at the plasma membrane. Because cathepsin proteases are activated by low pH in acidic endosomes, the acidification inhibitors may inhibit the viral infections by suppressing cathepsin protease activation. The acidification inhibitors attenuated the activities of cathepsin proteases B and L in TE671 cells, but not in XC cells. Processing of cathepsin protease L was suppressed by the acidification inhibitor in NIH3T3 cells, but again not in XC cells. These results indicate that cathepsin proteases are activated without endosome acidification in XC cells. Treatment with an endocytosis inhibitor or knockdown of dynamin 2 expression by siRNAs suppressed MLV infections in all examined cells including XC cells. Furthermore, endosomal cathepsin proteases were required for these viral infections in XC cells as other susceptible cells. These results suggest that infections of XC cells by the MLVs and Ebola virus occur through endosomes and pH-independent cathepsin activation induces pH-independent infection in XC cells.
Conflict of interest statement
Figures
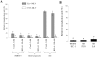








References
-
- Albritton LM, Tseng L, Scadden D, Cunningham JM. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

