Epigenetic virtues of chromodomains
- PMID: 22023491
- PMCID: PMC3223283
- DOI: 10.3109/10409238.2011.619164
Epigenetic virtues of chromodomains
Abstract
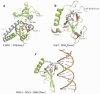
The chromatin organization modifier domain (chromodomain) was first identified as a motif associated with chromatin silencing in Drosophila. There is growing evidence that chromodomains are evolutionary conserved across different eukaryotic species to control diverse aspects of epigenetic regulation. Although originally reported as histone H3 methyllysine readers, the chromodomain functions have now expanded to recognition of other histone and non-histone partners as well as interaction with nucleic acids. Chromodomain binding to a diverse group of targets is mediated by a conserved substructure called the chromobox homology region. This motif can be used to predict methyllysine binding and distinguish chromodomains from related Tudor "Royal" family members. In this review, we discuss and classify various chromodomains according to their context, structure and the mechanism of target recognition.
Figures





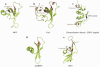

References
-
- AKHTAR A, BECKER PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–75. - PubMed
-
- AKHTAR A, ZINK D, BECKER PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–9. - PubMed
-
- ALLIS CD, BERGER SL, COTE J, DENT S, JENUWIEN T, KOUZARIDES T, PILLUS L, REINBERG D, SHI Y, SHIEKHATTAR R, SHILATIFARD A, WORKMAN J, ZHANG Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
