PNPASE and RNA trafficking into mitochondria
- PMID: 22023881
- PMCID: PMC3267854
- DOI: 10.1016/j.bbagrm.2011.10.001
PNPASE and RNA trafficking into mitochondria
Abstract
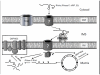
The mitochondrial genome encodes a very small fraction of the macromolecular components that are required to generate functional mitochondria. Therefore, most components are encoded within the nuclear genome and are imported into mitochondria from the cytosol. Understanding how mitochondria are assembled, function, and dysfunction in diseases requires detailed knowledge of mitochondrial import mechanisms and pathways. The import of nucleus-encoded RNAs is required for mitochondrial biogenesis and function, but unlike pre-protein import, the pathways and cellular machineries of RNA import are poorly defined, especially in mammals. Recent studies have shown that mammalian polynucleotide phosphorylase (PNPASE) localizes in the mitochondrial intermembrane space (IMS) to regulate the import of RNA. The identification of PNPASE as the first component of the RNA import pathway, along with a growing list of nucleus-encoded RNAs that are imported and newly developed assay systems for RNA import studies, suggest a unique opportunity is emerging to identify the factors and mechanisms that regulate RNA import into mammalian mitochondria. Here we summarize what is known in this fascinating area of mitochondrial biogenesis, identify areas that require further investigation, and speculate on the impact unraveling RNA import mechanisms and pathways will have for the field going forward. This article is part of a Special Issue entitled: Mitochondrial Gene Expression.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures



References
-
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. - PubMed
-
- Duchene AM, Pujol C, Marechal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Current genetics. 2009;55:1–18. - PubMed
-
- Sieber F, Duchene AM, Marechal-Drouard L. Mitochondrial RNA import: from diversity of natural mechanisms to potential applications. International review of cell and molecular biology. 2011;287:145–190. - PubMed
-
- Tarassov I, Kamenski P, Kolesnikova O, Karicheva O, Martin RP, Krasheninnikov IA, Entelis N. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell cycle (Georgetown, Tex. 2007;6:2473–2477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

