Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction
- PMID: 22024743
- PMCID: PMC3327613
- DOI: 10.4161/auto.7.12.18027
Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction
Abstract
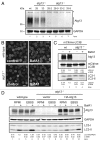
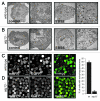
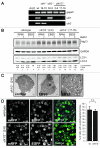
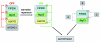
Under normal growth conditions the mammalian target of rapamycin complex 1 (mTORC1) negatively regulates the central autophagy regulator complex consisting of Unc-51-like kinases 1/2 (Ulk1/2), focal adhesion kinase family-interacting protein of 200 kDa (FIP200) and Atg13. Upon starvation, mTORC1-mediated repression of this complex is released, which then leads to Ulk1/2 activation. In this scenario, Atg13 has been proposed as an adaptor mediating the interaction between Ulk1/2 and FIP200 and enhancing Ulk1/2 kinase activity. Using Atg13-deficient cells, we demonstrate that Atg13 is indispensable for autophagy induction. We further show that Atg13 function strictly depends on FIP200 binding. In contrast, the simultaneous knockout of Ulk1 and Ulk2 did not have a similar effect on autophagy induction. Accordingly, the Ulk1-dependent phosphorylation sites we identified in Atg13 are expendable for this process. This suggests that Atg13 has an additional function independent of Ulk1/2 and that Atg13 and FIP200 act in concert during autophagy induction.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
