MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212
- PMID: 22024828
- PMCID: PMC3256021
- DOI: 10.1128/AAC.05270-11
MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212
Abstract
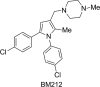
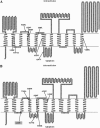
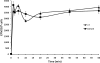
The 1,5-diarylpyrrole derivative BM212 was previously shown to be active against multidrug-resistant clinical isolates and Mycobacterium tuberculosis residing within macrophages as well as against Mycobacterium avium and other atypical mycobacteria. To determine its mechanism of action, we identified the cellular target. Spontaneous Mycobacterium smegmatis, Mycobacterium bovis BCG, and M. tuberculosis H37Rv mutants that were resistant to BM212 were isolated. By the screening of genomic libraries and by whole-genome sequencing, we found that all the characterized mutants showed mutations in the mmpL3 gene, allowing us to conclude that resistance to BM212 maps to the MmpL3 protein, a member of the MmpL (mycobacterial membrane protein, large) family. Susceptibility was unaffected by the efflux pump inhibitors reserpine, carbonylcyanide m-chlorophenylhydrazone, and verapamil. Uptake/efflux experiments with [(14)C]BM212 demonstrated that resistance is not driven by the efflux of BM212. Together, these data strongly suggest that the MmpL3 protein is the cellular target of BM212.
Figures
References
-
- Ainsa JA, Martin C, Cabeza M, De la Cruz F, Mendiola MV. 1996. Construction of a family of Mycobacterium/Escherichia coli shuttle vectors derived from pAL5000 and pACYC184: their use for cloning an antibiotic-resistance gene from Mycobacterium fortuitum. Gene 176:23–26 - PubMed
-
- Banerjee A, et al. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230 - PubMed
-
- Biava M. 2002. BM212 and its derivatives as a new class of antimycobacterial active agents. Curr. Med. Chem. 9:1859–1869 - PubMed
-
- Biava M, et al. 2011. Developing pyrrole-derived antimycobacterial agents: a rational lead optimization approach. ChemMedChem 6:593–598 - PubMed
-
- Chan ED, Iseman MD. 2008. Multidrug-resistant and extensively drug-resistant tuberculosis: a review. Curr. Opin. Infect. Dis. 21:587–595 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

