Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin
- PMID: 22025148
- PMCID: PMC3236649
- DOI: 10.1200/JCO.2010.33.8715
Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin
Abstract
Purpose: Mutation in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1(R132MUT)) is frequent in low-grade diffuse gliomas and, within glioblastoma (GBM), has been proposed as a marker for GBMs that arise by transformation from lower-grade gliomas, regardless of clinical history. To determine how GBMs arising with IDH1(R132MUT) differ from other GBMs, we undertook a comprehensive comparison of patients presenting clinically with primary GBM as a function of IDH1(R132) mutation status.
Patients and methods: In all, 618 treatment-naive primary GBMs and 235 lower-grade diffuse gliomas were sequenced for IDH1(R132) and analyzed for demographic, radiographic, anatomic, histologic, genomic, epigenetic, and transcriptional characteristics.
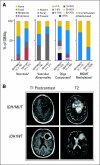
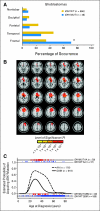
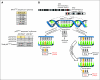
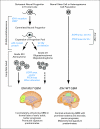
Results: Investigation revealed a constellation of features that distinguishes IDH1(R132MUT) GBMs from other GBMs (including frontal location and lesser extent of contrast enhancement and necrosis), relates them to lower-grade IDH1(R132MUT) gliomas, and supports the concept that IDH1(R132MUT) gliomas arise from a neural precursor population that is spatially and temporally restricted in the brain. The observed patterns of DNA sequence, methylation, and copy number alterations support a model of ordered molecular evolution of IDH1(R132MUT) GBM in which the appearance of mutant IDH1 protein is an initial event, followed by production of p53 mutant protein, and finally by copy number alterations of PTEN and EGFR.
Conclusion: Although histologically similar, GBMs arising with and without IDH1(R132MUT) appear to represent distinct disease entities that arise from separate cell types of origin as the result of largely nonoverlapping sets of molecular events. Optimal clinical management should account for the distinction between these GBM disease subtypes.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures





Comment in
-
Virchow 2011 or how to ID(H) human glioblastoma.J Clin Oncol. 2011 Dec 1;29(34):4473-4. doi: 10.1200/JCO.2011.37.5873. Epub 2011 Oct 24. J Clin Oncol. 2011. PMID: 22025161 No abstract available.
References
-
- Clarke J, Butowski N, Chang S. Recent advances in therapy for glioblastoma. Arch Neurol. 2010;67:279–283. - PubMed
-
- Hambardzumyan D, Squatrito M, Carbajal E, et al. Glioma formation, cancer stem cells, and akt signaling. Stem Cell Rev. 2008;4:203–210. - PubMed
-
- Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: A population-based study. Cancer Res. 2004;64:6892–6899. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

