Role of pro-oncogenic protein disulfide isomerase (PDI) family member anterior gradient 2 (AGR2) in the control of endoplasmic reticulum homeostasis
- PMID: 22025610
- PMCID: PMC3248018
- DOI: 10.1074/jbc.M111.275529
Role of pro-oncogenic protein disulfide isomerase (PDI) family member anterior gradient 2 (AGR2) in the control of endoplasmic reticulum homeostasis
Abstract
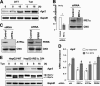
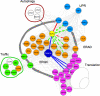
The protein-disulfide isomerase (PDI) family member anterior gradient 2 (AGR2) is reportedly overexpressed in numerous cancers and plays a role in cancer development. However, to date the molecular functions of AGR2 remain to be characterized. Herein we have identified AGR2 as bound to newly synthesized cargo proteins using a proteomics analysis of endoplasmic reticulum (ER) membrane-bound ribosomes. Nascent protein chains that translocate into the ER associate with specific ER luminal proteins, which in turn ensures proper folding and posttranslational modifications. Using both imaging and biochemical approaches, we confirmed that AGR2 localizes to the lumen of the ER and indirectly associates with ER membrane-bound ribosomes through nascent protein chains. We showed that AGR2 expression is controlled by the unfolded protein response and is in turn is involved in the maintenance of ER homeostasis. Remarkably, we have demonstrated that siRNA-mediated knockdown of AGR2 significantly alters the expression of components of the ER-associated degradation machinery and reduces the ability of cells to cope with acute ER stress, properties that might be relevant to the role of AGR2 in cancer development.
Figures







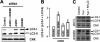
References
-
- Chevet E., Cameron P. H., Pelletier M. F., Thomas D. Y., Bergeron J. J. (2001) Curr. Opin. Struct. Biol. 11, 120–124 - PubMed
-
- Mori K. (2000) Cell 101, 451–454 - PubMed
-
- Kleizen B., Braakman I. (2004) Curr. Opin. Cell Biol. 16, 343–349 - PubMed
-
- Ruddock L. W., Molinari M. (2006) J. Cell Sci. 119, 4373–4380 - PubMed
-
- Määttänen P., Gehring K., Bergeron J. J., Thomas D. Y. (2010) Semin. Cell Dev. Biol. 21, 500–511 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

