CLUMPED CHLOROPLASTS 1 is required for plastid separation in Arabidopsis
- PMID: 22025705
- PMCID: PMC3215037
- DOI: 10.1073/pnas.1106706108
CLUMPED CHLOROPLASTS 1 is required for plastid separation in Arabidopsis
Abstract
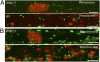
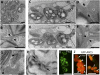
We identified an Arabidopsis thaliana mutant, clumped chloroplasts 1 (clmp1), in which disruption of a gene of unknown function causes chloroplasts to cluster instead of being distributed throughout the cytoplasm. The phenotype affects chloroplasts and nongreen plastids in multiple organs and cell types, but is detectable only at certain developmental stages. In young leaf petioles of clmp1, where clustering is prevalent, cells lacking chloroplasts are detected, suggesting impaired chloroplast partitioning during mitosis. Although organelle distribution and partitioning are actin-dependent in plants, the actin cytoskeleton in clmp1 is indistinguishable from that in WT, and peroxisomes and mitochondria are distributed normally. A CLMP1-YFP fusion protein that complements clmp1 localizes to discrete foci in the cytoplasm, most of which colocalize with the cell periphery or with chloroplasts. Ultrastructural analysis revealed that chloroplasts within clmp1 clusters are held together by membranous connections, including thin isthmi characteristic of late-stage chloroplast division. This finding suggests that constriction of dividing chloroplasts proceeds normally in clmp1, but separation is impaired. Consistently, chloroplast size and number, as well as positioning of the plastid division proteins FtsZ and ARC5/DRP5B, are unaffected in clmp1, indicating that loss of CLMP1-mediated chloroplast separation does not prevent otherwise normal division. CLMP1-like sequences are unique to green algae and land plants, and the CLMP1 sequence suggests that it functions through protein-protein interactions. Our studies identify a unique class of proteins required for plastid separation after the constriction stage of plastid division and indicate that CLMP1 activity is also required for plastid distribution and partitioning during cell division.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Wada M, Suetsugu N. Plant organelle positioning. Curr Opin Plant Biol. 2004;7:626–631. - PubMed
-
- Miyagishima SY, Kabeya Y. Chloroplast division: Squeezing the photosynthetic captive. Curr Opin Microbiol. 2010;13:738–746. - PubMed
-
- Osteryoung KW, Nunnari J. The division of endosymbiotic organelles. Science. 2003;302:1698–1704. - PubMed
-
- Kuroiwa T, et al. Vesicle, mitochondrial, and plastid division machineries with emphasis on dynamin and electron-dense rings. Int Rev Cell Mol Biol. 2008;271:97–152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

