Sortase enzymes in Gram-positive bacteria
- PMID: 22026821
- PMCID: PMC3590066
- DOI: 10.1111/j.1365-2958.2011.07887.x
Sortase enzymes in Gram-positive bacteria
Abstract
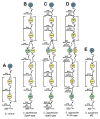
In Gram-positive bacteria proteins are displayed on the cell surface using sortase enzymes. These cysteine transpeptidases join proteins bearing an appropriate sorting signal to strategically positioned amino groups on the cell surface. Working alone, or in concert with other enzymes, sortases either attach proteins to the cross-bridge peptide of the cell wall or they link proteins together to form pili. Because surface proteins play a fundamental role in microbial physiology and are frequently virulence factors, sortase enzymes have been intensely studied since their discovery a little more than a decade ago. Based on their primary sequences and functions sortases can be partitioned into distinct families called class A to F enzymes. Most bacteria elaborate their surfaces using more than one type of sortase that function non-redundantly by recognizing unique sorting signals within their protein substrates. Here we review what is known about the functions of these enzymes and the molecular basis of catalysis. Particular emphasis is placed on 'pilin' specific class C sortases that construct structurally complex pili. Exciting new data have revealed that these enzymes are amazingly promiscuous in the substrates that they can employ and that there is a startling degree of diversity in their mechanism of action. We also review recent data that suggest that sortases are targeted to specific sites on the cell surface where they work with other sortases and accessory factors to properly function.
© 2011 Blackwell Publishing Ltd.
Figures

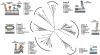

References
-
- Bentley ML, Gaweska H, Kielec JM, McCafferty DG. Engineering the substrate specificity of Staphylococcus aureus Sortase A. The beta6/beta7 loop from SrtB confers NPQTN recognition to SrtA. J Biol Chem. 2007;282:6571–6581. - PubMed
-
- Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66:495–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

