Generating social network data using partially described networks: an example informing avian influenza control in the British poultry industry
- PMID: 22027039
- PMCID: PMC3275467
- DOI: 10.1186/1746-6148-7-66
Generating social network data using partially described networks: an example informing avian influenza control in the British poultry industry
Abstract
Background: Targeted sampling can capture the characteristics of more vulnerable sectors of a population, but may bias the picture of population level disease risk. When sampling network data, an incomplete description of the population may arise leading to biased estimates of between-host connectivity. Avian influenza (AI) control planning in Great Britain (GB) provides one example where network data for the poultry industry (the Poultry Network Database or PND), targeted large premises and is consequently demographically biased. Exposing the effect of such biases on the geographical distribution of network properties could help target future poultry network data collection exercises. These data will be important for informing the control of potential future disease outbreaks.
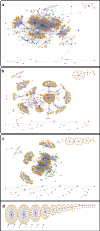
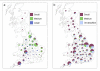
Results: The PND was used to compute between-farm association frequencies, assuming that farms sharing the same slaughterhouse or catching company, or through integration, are potentially epidemiologically linked. The fitted statistical models were extrapolated to the Great Britain Poultry Register (GBPR); this dataset is more representative of the poultry industry but lacks network information. This comparison showed how systematic biases in the demographic characterisation of a network, resulting from targeted sampling procedures, can bias the derived picture of between-host connectivity within the network.
Conclusions: With particular reference to the predictive modeling of AI in GB, we find significantly different connectivity patterns across GB when network estimates incorporate the more demographically representative information provided by the GBPR; this has not been accounted for by previous epidemiological analyses. We recommend ranking geographical regions, based on relative confidence in extrapolated estimates, for prioritising further data collection. Evaluating whether and how the between-farm association frequencies impact on the risk of between-farm transmission will be the focus of future work.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

