Dimerization of oskar 3' UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte
- PMID: 22028360
- PMCID: PMC3222118
- DOI: 10.1261/rna.2686411
Dimerization of oskar 3' UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte
Abstract
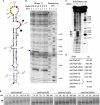
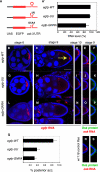
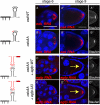
mRNA localization coupled with translational control is a highly conserved and widespread mechanism for restricting protein expression to specific sites within eukaryotic cells. In Drosophila, patterning of the embryo requires oskar mRNA transport to the posterior pole of the oocyte and translational repression prior to localization. oskar RNA splicing and the 3' untranslated region (UTR) are required for posterior enrichment of the mRNA. However, reporter RNAs harboring the oskar 3' UTR can localize by hitchhiking with endogenous oskar transcripts. Here we show that the oskar 3' UTR contains a stem-loop structure that promotes RNA dimerization in vitro and hitchhiking in vivo. Mutations in the loop that abolish in vitro dimerization interfere with reporter RNA localization, and restoring loop complementarity restores hitchhiking. Our analysis provides insight into the molecular basis of RNA hitchhiking, whereby localization-incompetent RNA molecules can become locally enriched in the cytoplasm, by virtue of their association with transport-competent RNAs.
Figures

References
-
- Brunel C, Marquet R, Romby P, Ehresmann C 2002. RNA loop–loop interactions as dynamic functional motifs. Biochimie 84: 925–944 - PubMed
-
- Chekulaeva M, Hentze MW, Ephrussi A 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124: 521–533 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
