Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination
- PMID: 22028627
- PMCID: PMC3196474
- DOI: 10.1371/journal.pbio.1001176
Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination
Abstract
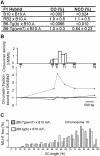
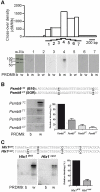
Meiotic recombination generates reciprocal exchanges between homologous chromosomes (also called crossovers, COs) that are essential for proper chromosome segregation during meiosis and are a major source of genome diversity by generating new allele combinations. COs have two striking properties: they occur at specific sites, called hotspots, and these sites evolve rapidly. In mammals, the Prdm9 gene, which encodes a meiosis-specific histone H3 methyltransferase, has recently been identified as a determinant of CO hotspots. Here, using transgenic mice, we show that the sole modification of PRDM9 zinc fingers leads to changes in hotspot activity, histone H3 lysine 4 trimethylation (H3K4me3) levels, and chromosome-wide distribution of COs. We further demonstrate by an in vitro assay that the PRDM9 variant associated with hotspot activity binds specifically to DNA sequences located at the center of the three hotspots tested. Remarkably, we show that mutations in cis located at hotspot centers and associated with a decrease of hotspot activity affect PRDM9 binding. Taken together, these results provide the direct demonstration that Prdm9 is a master regulator of hotspot localization through the DNA binding specificity of its zinc finger array and that binding of PRDM9 at hotspots promotes local H3K4me3 enrichment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–399. - PubMed
-
- Buard J, de Massy B. Playing hide and seek with mammalian meiotic crossover hotspots. Trends Genet. 2007;23:301–309. - PubMed
-
- Jeffreys A. J, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. - PubMed
-
- Jeffreys A. J, Neumann R, Panayi M, Myers S, Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nat Genet. 2005;37:601–606. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

