Most random gene expression signatures are significantly associated with breast cancer outcome
- PMID: 22028643
- PMCID: PMC3197658
- DOI: 10.1371/journal.pcbi.1002240
Most random gene expression signatures are significantly associated with breast cancer outcome
Abstract
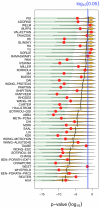
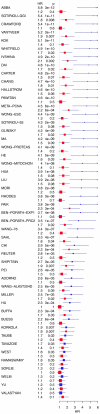
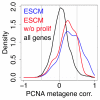
Bridging the gap between animal or in vitro models and human disease is essential in medical research. Researchers often suggest that a biological mechanism is relevant to human cancer from the statistical association of a gene expression marker (a signature) of this mechanism, that was discovered in an experimental system, with disease outcome in humans. We examined this argument for breast cancer. Surprisingly, we found that gene expression signatures-unrelated to cancer-of the effect of postprandial laughter, of mice social defeat and of skin fibroblast localization were all significantly associated with breast cancer outcome. We next compared 47 published breast cancer outcome signatures to signatures made of random genes. Twenty-eight of them (60%) were not significantly better outcome predictors than random signatures of identical size and 11 (23%) were worst predictors than the median random signature. More than 90% of random signatures >100 genes were significant outcome predictors. We next derived a metagene, called meta-PCNA, by selecting the 1% genes most positively correlated with proliferation marker PCNA in a compendium of normal tissues expression. Adjusting breast cancer expression data for meta-PCNA abrogated almost entirely the outcome association of published and random signatures. We also found that, in the absence of adjustment, the hazard ratio of outcome association of a signature strongly correlated with meta-PCNA (R(2) = 0.9). This relation also applied to single-gene expression markers. Moreover, >50% of the breast cancer transcriptome was correlated with meta-PCNA. A corollary was that purging cell cycle genes out of a signature failed to rule out the confounding effect of proliferation. Hence, it is questionable to suggest that a mechanism is relevant to human breast cancer from the finding that a gene expression marker for this mechanism predicts human breast cancer outcome, because most markers do. The methods we present help to overcome this problem.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. - DOI - PubMed
-
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AAM, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. - DOI - PubMed
-
- Paik S, Shak S, Tang G, Kim C, Baker J, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. - DOI - PubMed
-
- Pawitan Y, Bjöhle J, Amler L, Borg A-L, Egyhazi S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. - DOI - PMC - PubMed
-
- Korkola JE, Blaveri E, DeVries S, Moore DH, Hwang ES, et al. Identification of a robust gene signature that predicts breast cancer outcome in independent data sets. BMC Cancer. 2007;7:61. doi: 10.1186/1471-2407-7-61. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

