A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum
- PMID: 22028654
- PMCID: PMC3197617
- DOI: 10.1371/journal.ppat.1002310
A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum
Abstract
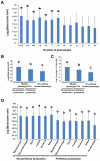
Fusarium graminearum is an important plant pathogen that causes head blight of major cereal crops. The fungus produces mycotoxins that are harmful to animal and human. In this study, a systematic analysis of 17 phenotypes of the mutants in 657 Fusarium graminearum genes encoding putative transcription factors (TFs) resulted in a database of over 11,000 phenotypes (phenome). This database provides comprehensive insights into how this cereal pathogen of global significance regulates traits important for growth, development, stress response, pathogenesis, and toxin production and how transcriptional regulations of these traits are interconnected. In-depth analysis of TFs involved in sexual development revealed that mutations causing defects in perithecia development frequently affect multiple other phenotypes, and the TFs associated with sexual development tend to be highly conserved in the fungal kingdom. Besides providing many new insights into understanding the function of F. graminearum TFs, this mutant library and phenome will be a valuable resource for characterizing the gene expression network in this fungus and serve as a reference for studying how different fungi have evolved to control various cellular processes at the transcriptional level.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Shelest E. Transcription factors in fungi. FEMS Microbiol Lett. 2008;286:145–151. - PubMed
-
- Leslie JF, Summerell BA. The Fusarium laboratory manual. Ames, IA: Blackwell Pub; 2006.
-
- Desjardins AE, Proctor RH. Molecular biology of Fusarium mycotoxins. Int J Food Microbiol. 2007;119:47–50. - PubMed
-
- Cuomo CA, Guldener U, Xu J-R, Trail F, Turgeon BG, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. - PubMed
-
- Hallen HE, Huebner M, Shiu S-H, Güldener U, Trail F. Gene expression shifts during perithecium development in Gibberella zeae (anamorph Fusarium graminearum), with particular emphasis on ion transport proteins. Fungal Genet Biol. 2007;44:1146–1156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

