A J-like protein influences fatty acid composition of chloroplast lipids in Arabidopsis
- PMID: 22028775
- PMCID: PMC3196505
- DOI: 10.1371/journal.pone.0025368
A J-like protein influences fatty acid composition of chloroplast lipids in Arabidopsis
Abstract
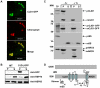
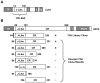
A comprehensive understanding of the lipid and fatty acid metabolic machinery is needed for optimizing production of oils and fatty acids for fuel, industrial feedstocks and nutritional improvement in plants. T-DNA mutants in the poorly annotated Arabidopsis thaliana gene At1g08640 were identified as containing moderately high levels (50-100%) of 16∶1Δ7 and 18∶1Δ9 leaf fatty acids and subtle decreases (5-30%) of 16∶3 and 18∶3 (http://www.plastid.msu.edu/). TLC separation of fatty acids in the leaf polar lipids revealed that the chloroplastic galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) were the main lipid types affected by this mutation. Analysis of the inferred amino acid sequence of At1g08640 predicted the presence of a transit peptide, three transmembrane domains and an N-terminal J-like domain, and the gene was named CJD1 for Chloroplast J-like Domain 1. GFP reporter experiments and in vitro chloroplast import assays demonstrated CJD1 is a chloroplast membrane protein. Screening of an Arabidopsis cDNA library by yeast-2-hybrid (Y2H) using the J-like domain of CJD1 as bait identified a plastidial inner envelope protein (Accumulation and Replication of Chloroplasts 6, ARC6) as the primary interacting partner in the Y2H assay. ARC6 plays a central role in chloroplast division and binds CJD1 via its own J-like domain along with an adjacent conserved region whose function is not fully known. These results provide a starting point for future investigations of how mutations in CJD1 affect lipid composition.
Conflict of interest statement
Figures



References
-
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14:354–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

