The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer
- PMID: 22028892
- PMCID: PMC3197668
- DOI: 10.1371/journal.pone.0026514
The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer
Abstract
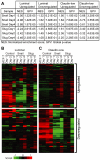
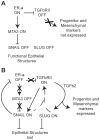
The transcriptional repressors Snail and Slug are situated at the core of several signaling pathways proposed to mediate epithelial to mesenchymal transition or EMT, which has been implicated in tumor metastasis. EMT involves an alteration from an organized, epithelial cell structure to a mesenchymal, invasive and migratory phenotype. In order to obtain a global view of the impact of Snail and Slug expression, we performed a microarray experiment using the MCF-7 breast cancer cell line, which does not express detectable levels of Snail or Slug. MCF-7 cells were infected with Snail, Slug or control adenovirus, and RNA samples isolated at various time points were analyzed across all transcripts. Our analyses indicated that Snail and Slug regulate many genes in common, but also have distinct sets of gene targets. Gene set enrichment analyses indicated that Snail and Slug directed the transcriptome of MCF-7 cells from a luminal towards a more complex pattern that includes many features of the claudin-low breast cancer signature. Of particular interest, genes involved in the TGF-beta signaling pathway are upregulated, while genes responsible for a differentiated morphology are downregulated following Snail or Slug expression. Further we noticed increased histone acetylation at the promoter region of the transforming growth factor beta-receptor II (TGFBR2) gene following Snail or Slug expression. Inhibition of the TGF-beta signaling pathway using selective small-molecule inhibitors following Snail or Slug addition resulted in decreased cell migration with no impact on the repression of cell junction molecules by Snail and Slug. We propose that there are two regulatory modules embedded within EMT: one that involves repression of cell junction molecules, and the other involving cell migration via TGF-beta and/or other pathways.
Conflict of interest statement
Figures




References
-
- Blick T, Hugo H, Widodo E, Waltham M, Pinto C, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:235–252. - PubMed
-
- Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–945. - PubMed
-
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. - PubMed
-
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

