Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding
- PMID: 22028919
- PMCID: PMC3197680
- DOI: 10.1371/journal.pone.0026601
Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding
Abstract
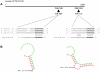
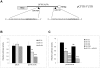
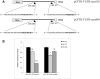
microRNAs (miRNAs) are a class of regulatory small non-coding molecules that control gene expression at post-transcriptional level. Deregulation of miRNA functions affects a variety of biological processes also involved in the etiology of several human mendelian and complex diseases. Recently, aberrant miRNA expression has been observed in Cystic Fibrosis (CF), an autosomal-recessive genetic disorder caused by mutations in the CFTR gene, in which a genotype-phenotype correlation is not always found. In order to determine miRNA role in CFTR post-transcriptional regulation, we searched for miR-responsive elements in the CFTR 3'-UTR. In silico analysis, performed using different computational on-line programs, identified some putative miRNAs. Both miR-101 and miR-494 synthetic mimics significantly inhibited the expression of a reporter construct containing the 3'-UTR of CFTR in luciferase assays. Interestingly, miR-101/miR-494 combination was able to markedly suppress CFTR activity by approximately 80% (p<0.001). This is one of the first in vitro studies implicating microRNAs as negative regulators of the CFTR gene expression. miRNA aberrant expression and function might explain the wide phenotypic variability observed among CF patients.
Conflict of interest statement
Figures
References
-
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. - PubMed
-
- Xu W, Hui C, Yu SS, Jing C, Chan HC. MicroRNAs and cystic fibrosis–an epigenetic perspective. Cell Biol Int. 2011;35:463–466. - PubMed
-
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

