Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait
- PMID: 22031822
- PMCID: PMC3198726
- DOI: 10.1371/journal.pone.0026147
Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait
Abstract
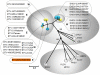
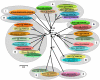
Bluetongue virus is the "type" species of the genus Orbivirus, family Reoviridae. Twenty four distinct bluetongue virus (BTV) serotypes have been recognized for decades, any of which is thought to be capable of causing "bluetongue" (BT), an insect-borne disease of ruminants. However, two further BTV serotypes, BTV-25 (Toggenburg orbivirus, from Switzerland) and BTV-26 (from Kuwait) have recently been identified in goats and sheep, respectively. The BTV genome is composed of ten segments of linear dsRNA, encoding 7 virus-structural proteins (VP1 to VP7) and four distinct non-structural (NS) proteins (NS1 to NS4). We report the entire BTV-26 genome sequence (isolate KUW2010/02) and comparisons to other orbiviruses. Highest identity levels were consistently detected with other BTV strains, identifying KUW2010/02 as BTV. The outer-core protein and major BTV serogroup-specific antigen "VP7" showed 98% aa sequence identity with BTV-25, indicating a common ancestry. However, higher level of variation in the nucleotide sequence of Seg-7 (81.2% identity) suggests strong conservation pressures on the protein of these two strains, and that they diverged a long time ago. Comparisons of Seg-2, encoding major outer-capsid component and cell-attachment protein "VP2" identified KUW2010/02 as 26th BTV, within a 12th Seg-2 nucleotype [nucleotype L]. Comparisons of Seg-6, encoding the smaller outer capsid protein VP5, also showed levels of nt/aa variation consistent with identification of KUW2010/02 as BTV-26 (within a 9th Seg-6 nucleotype - nucleotype I). Sequence data for Seg-2 of KUW2010/02 were used to design four sets of oligonucleotide primers for use in BTV-26, type-specific RT-PCR assays. Analyses of other more conserved genome segments placed KUW2010/02 and BTV-25/SWI2008/01 closer to each other than to other "eastern" or "western" BTV strains, but as representatives of two novel and distinct geographic groups (topotypes). Our analyses indicate that all of the BTV genome segments have evolved under strong purifying selection.
Conflict of interest statement
Figures

References
-
- Mertens PPC, Maan S, Samuel A, Attoui H. Orbiviruses, Reoviridae. In: Fauquet CM, Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy Eighth Report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2005. pp. 466–483.
-
- Attoui H, Maan SS, Anthony SJ, Mertens PPC, editors. 1 ed. London: Elsevier/Academic Press; 2009. Bluetongue virus, other orbiviruses and other reoviruses: Their relationships and taxonomy. pp. 23–552.
-
- Alexander KA, MacLachlan NJ, Kat PW, House C, O'Brien SJ, et al. Evidence of natural bluetongue virus infection among African carnivores. Am J Trop Med Hyg. 1994;51:568–576. - PubMed
-
- Meyer G, Lacroux C, Leger S, Top S, Goyeau K, et al. Lethal bluetongue virus serotype 1 infection in llamas. Emerg Infect Dis. 2009;15:608–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

