Protein dynamics and enzymatic chemical barrier passage
- PMID: 22031954
- PMCID: PMC3245361
- DOI: 10.1021/jp207876k
Protein dynamics and enzymatic chemical barrier passage
Abstract
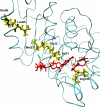
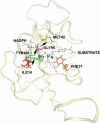
After many decades of investigation, the manner in which enzymes increase the rate of chemical reactions, at times by a factor of 10(17) compared to the rate of the corresponding solution phase reaction, is still opaque. A topic of significant discussion in the literature of the past 5-10 years has been the importance of protein dynamics in this process. This Feature Article will discuss the authors' work on this still controversial topic with focus on both methodology and application to real systems. The end conclusion of this work has been that for specific enzymes under study protein dynamics on both rapid time scales of barrier crossing (termed promoting vibrations by the authors) and of conformational fluctuations are central to the function of biological catalysts. In another enzyme we will discuss, the results are far less clear. The manner of the coupling of chemistry to protein dynamics has deep implications for protein architecture, both natural and created, and recent results reinforce the complexity of the protein form that has evolved to support these functions.
© 2011 American Chemical Society
Figures

References
-
- Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE. Science. 2011;332:234–8. - PMC - PubMed
- Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Nature. 2009;462:669–673. - PMC - PubMed
- Boehr DD, McElheny D, Dyson HJ, Wright PE. Proc. Natl. Acad. Sci. USA. 2010;107:1373–8. - PMC - PubMed
-
- Ghanem M, Li L, Wing C, Schramm VL. Biochem. 2008;47:2559–2564. - PubMed
-
- Kramers HA. Physica. 1940;4:284.
-
- Zwanzig R. J. Stat. Phys. 1973;9:215.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

