Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles
- PMID: 22032172
- PMCID: PMC3333779
- DOI: 10.1186/gb-2011-12-10-r109
Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles
Abstract
Background: It is widely acknowledged that synonymous codons are used unevenly among genes in a genome. In organisms under translational selection, genes encoding highly expressed proteins are enriched with specific codons. This phenomenon, termed codon usage bias, is common to many organisms and has been recognized as influencing cellular fitness. This suggests that the global extent of codon usage bias of an organism might be associated with its phenotypic traits.
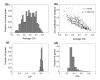
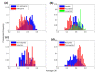
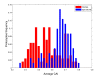
Results: To test this hypothesis we used a simple measure for assessing the extent of codon bias of an organism, and applied it to hundreds of sequenced prokaryotes. Our analysis revealed a large variability in this measure: there are organisms showing very high degrees of codon usage bias and organisms exhibiting almost no differential use of synonymous codons among different genes. Remarkably, we found that the extent of codon usage bias corresponds to the lifestyle of the organism. Especially, organisms able to live in a wide range of habitats exhibit high extents of codon usage bias, consistent with their need to adapt efficiently to different environments. Pathogenic prokaryotes also demonstrate higher extents of codon usage bias than non-pathogenic prokaryotes, in accord with the multiple environments that many pathogens occupy. Our results show that the previously observed correlation between growth rate and metabolic variability is attributed to their individual associations with codon usage bias.
Conclusions: Our results suggest that the extent of codon usage bias of an organism plays a role in the adaptation of prokaryotes to their environments.
Figures
References
-
- Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. - PubMed
-
- Post LE, Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem. 1980;255:4660–4666. - PubMed
-
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

