Automated NMR relaxation dispersion data analysis using NESSY
- PMID: 22032230
- PMCID: PMC3215250
- DOI: 10.1186/1471-2105-12-421
Automated NMR relaxation dispersion data analysis using NESSY
Abstract
Background: Proteins are dynamic molecules with motions ranging from picoseconds to longer than seconds. Many protein functions, however, appear to occur on the micro to millisecond timescale and therefore there has been intense research of the importance of these motions in catalysis and molecular interactions. Nuclear Magnetic Resonance (NMR) relaxation dispersion experiments are used to measure motion of discrete nuclei within the micro to millisecond timescale. Information about conformational/chemical exchange, populations of exchanging states and chemical shift differences are extracted from these experiments. To ensure these parameters are correctly extracted, accurate and careful analysis of these experiments is necessary.
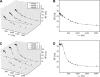
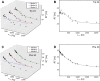
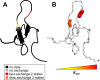
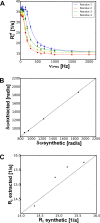
Results: The software introduced in this article is designed for the automatic analysis of relaxation dispersion data and the extraction of the parameters mentioned above. It is written in Python for multi platform use and highest performance. Experimental data can be fitted to different models using the Levenberg-Marquardt minimization algorithm and different statistical tests can be used to select the best model. To demonstrate the functionality of this program, synthetic data as well as NMR data were analyzed. Analysis of these data including the generation of plots and color coded structures can be performed with minimal user intervention and using standard procedures that are included in the program.
Conclusions: NESSY is easy to use open source software to analyze NMR relaxation data. The robustness and standard procedures are demonstrated in this article.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

