Identification of hunchback cis-regulatory DNA conferring temporal expression in neuroblasts and neurons
- PMID: 22033538
- PMCID: PMC3272097
- DOI: 10.1016/j.gep.2011.10.001
Identification of hunchback cis-regulatory DNA conferring temporal expression in neuroblasts and neurons
Abstract
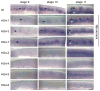
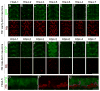
The specification of temporal identity within single progenitor lineages is essential to generate functional neuronal diversity in Drosophila and mammals. In Drosophila, four transcription factors are sequentially expressed in neural progenitors (neuroblasts) and each regulates the temporal identity of the progeny produced during its expression window. The first temporal identity is established by the Ikaros-family zinc finger transcription factor Hunchback (Hb). Hb is detected in young (newly-formed) neuroblasts for about an hour and is maintained in the early-born neurons produced during this interval. Hb is necessary and sufficient to specify early-born neuronal or glial identity in multiple neuroblast lineages. The timing of hb expression in neuroblasts is regulated at the transcriptional level. Here we identify cis-regulatory elements that confer proper hb expression in "young" neuroblasts and early-born neurons. We show that the neuroblast element contains clusters of predicted binding sites for the Seven-up transcription factor, which is known to limit hb neuroblast expression. We identify highly conserved sequences in the neuronal element that are good candidates for maintaining Hb transcription in neurons. Our results provide the necessary foundation for identifying trans-acting factors that establish the Hb early temporal expression domain.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures





References
-
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226:34–44. - PubMed
-
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. - PubMed
-
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. - PubMed
-
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

