A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans
- PMID: 22035958
- PMCID: PMC3384509
- DOI: 10.1016/j.cell.2011.09.045
A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans
Abstract
Sterol regulatory element-binding proteins (SREBPs) activate genes involved in the synthesis and trafficking of cholesterol and other lipids and are critical for maintaining lipid homeostasis. Aberrant SREBP activity, however, can contribute to obesity, fatty liver disease, and insulin resistance, hallmarks of metabolic syndrome. Our studies identify a conserved regulatory circuit in which SREBP-1 controls genes in the one-carbon cycle, which produces the methyl donor S-adenosylmethionine (SAMe). Methylation is critical for the synthesis of phosphatidylcholine (PC), a major membrane component, and we find that blocking SAMe or PC synthesis in C. elegans, mouse liver, and human cells causes elevated SREBP-1-dependent transcription and lipid droplet accumulation. Distinct from negative regulation of SREBP-2 by cholesterol, our data suggest a feedback mechanism whereby maturation of nuclear, transcriptionally active SREBP-1 is controlled by levels of PC. Thus, nutritional or genetic conditions limiting SAMe or PC production may activate SREBP-1, contributing to human metabolic disorders.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
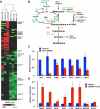
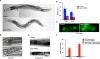
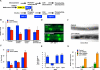
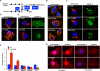

Comment in
-
New driver for lipid synthesis.Cell. 2011 Nov 11;147(4):719-21. doi: 10.1016/j.cell.2011.10.019. Cell. 2011. PMID: 22078872
References
-
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. - PubMed
-
- Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. - PubMed
-
- Brendza KM, Haakenson W, Cahoon RE, Hicks LM, Palavalli LH, Chiapelli BJ, McLaird M, McCarter JP, Williams DJ, Hresko MC, et al. Phosphoethanolamine N-methyltransferase (PMT-1) catalyses the first reaction of a new pathway for phosphocholine biosynthesis in Caenorhabditis elegans. Biochemical Journal. 2007;404:439–448. - PMC - PubMed
-
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

