The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma
- PMID: 22036564
- PMCID: PMC3208379
- DOI: 10.1016/j.cell.2011.09.035
The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma
Abstract
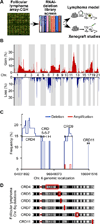
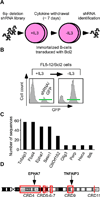
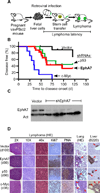
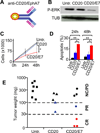
Insights into cancer genetics can lead to therapeutic opportunities. By cross-referencing chromosomal changes with an unbiased genetic screen we identify the ephrin receptor A7 (EPHA7) as a tumor suppressor in follicular lymphoma (FL). EPHA7 is a target of 6q deletions and inactivated in 72% of FLs. Knockdown of EPHA7 drives lymphoma development in a murine FL model. In analogy to its physiological function in brain development, a soluble splice variant of EPHA7 (EPHA7(TR)) interferes with another Eph-receptor and blocks oncogenic signals in lymphoma cells. Consistent with this drug-like activity, administration of the purified EPHA7(TR) protein produces antitumor effects against xenografted human lymphomas. Further, by fusing EPHA7(TR) to the anti-CD20 antibody (rituximab) we can directly target this tumor suppressor to lymphomas in vivo. Our study attests to the power of combining descriptive tumor genomics with functional screens and reveals EPHA7(TR) as tumor suppressor with immediate therapeutic potential.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Lymphoma. Suppressive EPH-ect.Nat Rev Cancer. 2011 Nov 17;11(12):829. doi: 10.1038/nrc3178. Nat Rev Cancer. 2011. PMID: 22089418 No abstract available.
-
Discovery of a secreted tumor suppressor provides a promising therapeutic strategy for follicular lymphoma.Cancer Cell. 2011 Nov 15;20(5):559-61. doi: 10.1016/j.ccr.2011.10.027. Cancer Cell. 2011. PMID: 22094251
References
-
- Barr PM, Lazarus HM. Follicular non-Hodgkin lymphoma: long-term results of stem-cell transplantation. Curr Opin Oncol. 2008;20:502–508. - PubMed
-
- Bende RJ, Smit LA, van Noesel CJ. Molecular pathways in follicular lymphoma. Leukemia. 2007;21:18–29. - PubMed
-
- Calado DP, Zhang B, Srinivasan L, Sasaki Y, Seagal J, Unitt C, Rodig S, Kutok J, Tarakhovsky A, Schmidt-Supprian M, et al. Constitutive canonical NF-kappaB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18:580–589. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

