Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity
- PMID: 22037377
- PMCID: PMC3230241
- DOI: 10.1038/nbt.2017
Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity
Abstract
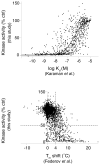
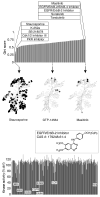
Small-molecule protein kinase inhibitors are widely used to elucidate cellular signaling pathways and are promising therapeutic agents. Owing to evolutionary conservation of the ATP-binding site, most kinase inhibitors that target this site promiscuously inhibit multiple kinases. Interpretation of experiments that use these compounds is confounded by a lack of data on the comprehensive kinase selectivity of most inhibitors. Here we used functional assays to profile the activity of 178 commercially available kinase inhibitors against a panel of 300 recombinant protein kinases. Quantitative analysis revealed complex and often unexpected interactions between protein kinases and kinase inhibitors, with a wide spectrum of promiscuity. Many off-target interactions occur with seemingly unrelated kinases, revealing how large-scale profiling can identify multitargeted inhibitors of specific, diverse kinases. The results have implications for drug development and provide a resource for selecting compounds to elucidate kinase function and for interpreting the results of experiments involving kinase inhibitors.
Conflict of interest statement
Figures



Comment in
-
Interrogating the kinome.Nat Biotechnol. 2011 Nov 8;29(11):981-3. doi: 10.1038/nbt.2021. Nat Biotechnol. 2011. PMID: 22068532 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

