SNPpy--database management for SNP data from genome wide association studies
- PMID: 22039405
- PMCID: PMC3198468
- DOI: 10.1371/journal.pone.0024982
SNPpy--database management for SNP data from genome wide association studies
Abstract
Background: We describe SNPpy, a hybrid script database system using the Python SQLAlchemy library coupled with the PostgreSQL database to manage genotype data from Genome-Wide Association Studies (GWAS). This system makes it possible to merge study data with HapMap data and merge across studies for meta-analyses, including data filtering based on the values of phenotype and Single-Nucleotide Polymorphism (SNP) data. SNPpy and its dependencies are open source software.
Results: The current version of SNPpy offers utility functions to import genotype and annotation data from two commercial platforms. We use these to import data from two GWAS studies and the HapMap Project. We then export these individual datasets to standard data format files that can be imported into statistical software for downstream analyses.
Conclusions: By leveraging the power of relational databases, SNPpy offers integrated management and manipulation of genotype and phenotype data from GWAS studies. The analysis of these studies requires merging across GWAS datasets as well as patient and marker selection. To this end, SNPpy enables the user to filter the data and output the results as standardized GWAS file formats. It does low level and flexible data validation, including validation of patient data. SNPpy is a practical and extensible solution for investigators who seek to deploy central management of their GWAS data.
Conflict of interest statement
Figures



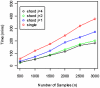
 and
and  . For all these datasets, the number of SNPs is 620,901.
. For all these datasets, the number of SNPs is 620,901.
 and Geno Shard layout with degree of parallelism
and Geno Shard layout with degree of parallelism  and
and  . For all these datasets, the number of SNPs is 620,901. All timings correspond to warm cache.
. For all these datasets, the number of SNPs is 620,901. All timings correspond to warm cache.
References
-
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. - PubMed
-
- GLU: Genotype Library and Utilities. URL http://code.google.com/p/glu-genetics/. Accessed: 2011 September 27th.
-
- Conway JE. PL/R: a PostgreSQL loadable procedural language handler for the R programming language. 2009. URL http://www.joeconway.com/plr/. Version 8.3.0.8. Accessed: 2011 September 27th.
-
- Wellcome Trust Case Control Consortium. URL https://www.wtccc.org.uk/info/access_to_data_samples.shtml. Accessed: 2011 September 27th.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

