FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection
- PMID: 22039434
- PMCID: PMC3198457
- DOI: 10.1371/journal.pone.0026047
FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection
Erratum in
- PLoS One. 2011;6(11). doi: 10.1371/annotation/468cfdcd-184c-42f7-a1d0-3b72a2f6a558
Abstract
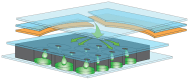
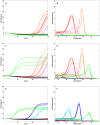
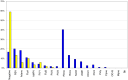
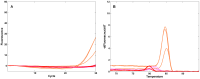
The ideal clinical diagnostic system should deliver rapid, sensitive, specific and reproducible results while minimizing the requirements for specialized laboratory facilities and skilled technicians. We describe an integrated diagnostic platform, the "FilmArray", which fully automates the detection and identification of multiple organisms from a single sample in about one hour. An unprocessed biologic/clinical sample is subjected to nucleic acid purification, reverse transcription, a high-order nested multiplex polymerase chain reaction and amplicon melt curve analysis. Biochemical reactions are enclosed in a disposable pouch, minimizing the PCR contamination risk. FilmArray has the potential to detect greater than 100 different nucleic acid targets at one time. These features make the system well-suited for molecular detection of infectious agents. Validation of the FilmArray technology was achieved through development of a panel of assays capable of identifying 21 common viral and bacterial respiratory pathogens. Initial testing of the system using both cultured organisms and clinical nasal aspirates obtained from children demonstrated an analytical and clinical sensitivity and specificity comparable to existing diagnostic platforms. We demonstrate that automated identification of pathogens from their corresponding target amplicon(s) can be accomplished by analysis of the DNA melting curve of the amplicon.
Conflict of interest statement
Figures


References
-
- Petric M, Comanor L, Petti CA. Role of the laboratory in diagnosis of influenza during seasonal epidemics and potential pandemics. J Infect Dis. 2006;194(Suppl 2):S98–110. - PubMed
-
- Lessa FC, Gould PL, Pascoe N, Erdman DD, Lu X, et al. Health care transmission of a newly emergent adenovirus serotype in health care personnel at a military hospital in Texas, 2007. J Infect Dis. 2009;200:1759–1765. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

